UniProt ID: Q9UHD9 (1-624) Ubiquilin-2 (R12G, R15D, Q21E)
|
|
|
|
| Sample: |
Ubiquilin-2 (R12G, R15D, Q21E) dimer, 131 kDa Homo sapiens protein
|
| Buffer: |
20 mM sodium phosphate, 0.5 mM EDTA, 0.02 % NaN3, pH: 6.8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2023 Mar 22
|
UBQLN2 domain deletion constructs
Sarasi Galagedera
|
| RgGuinier |
5.1 |
nm |
| Dmax |
21.3 |
nm |
| VolumePorod |
330 |
nm3 |
|
|
UniProt ID: Q9UHD9 (1-624) Ubiquilin-2 (∆551-569)
|
|
|
|
| Sample: |
Ubiquilin-2 (∆551-569) dimer, 127 kDa Homo sapiens protein
|
| Buffer: |
20 mM sodium phosphate, 0.5 mM EDTA, 0.02 % NaN3, pH: 6.8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2023 Mar 22
|
UBQLN2 domain deletion constructs
Sarasi Galagedera
|
| RgGuinier |
5.2 |
nm |
| Dmax |
23.4 |
nm |
| VolumePorod |
332 |
nm3 |
|
|
UniProt ID: P54727 (1-409) UV excision repair protein RAD23 homolog B
|
|
|
|
| Sample: |
UV excision repair protein RAD23 homolog B monomer, 43 kDa Homo sapiens protein
|
| Buffer: |
20 mM sodium phosphate, 0.5 mM EDTA, 0.02 % NaN3, pH: 6.8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Feb 22
|
Domain deletion constructs of hHR23B
Sarasi Galagedera
|
| RgGuinier |
4.6 |
nm |
| Dmax |
26.0 |
nm |
| VolumePorod |
103 |
nm3 |
|
|
UniProt ID: P54727 (80-409) UV excision repair protein RAD23 homolog B (∆1-79)
|
|
|
|
| Sample: |
UV excision repair protein RAD23 homolog B (∆1-79) monomer, 35 kDa Homo sapiens protein
|
| Buffer: |
20 mM sodium phosphate, 0.5 mM EDTA, 0.02 % NaN3, pH: 6.8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Feb 22
|
Domain deletion constructs of hHR23B
Sarasi Galagedera
|
| RgGuinier |
4.6 |
nm |
| Dmax |
21.6 |
nm |
| VolumePorod |
92 |
nm3 |
|
|
UniProt ID: P54727 (1-409) UV excision repair protein RAD23 homolog B (∆186-231)
|
|
|
|
| Sample: |
UV excision repair protein RAD23 homolog B (∆186-231) monomer, 38 kDa Homo sapiens protein
|
| Buffer: |
20 mM sodium phosphate, 0.5 mM EDTA, 0.02 % NaN3, pH: 6.8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Feb 22
|
Domain deletion constructs of hHR23B
Sarasi Galagedera
|
| RgGuinier |
4.6 |
nm |
| Dmax |
21.7 |
nm |
| VolumePorod |
95 |
nm3 |
|
|
UniProt ID: P54727 (1-361) UV excision repair protein RAD23 homolog B (∆362-409)
|
|
|
|
| Sample: |
UV excision repair protein RAD23 homolog B (∆362-409) monomer, 38 kDa Homo sapiens protein
|
| Buffer: |
20 mM sodium phosphate, 0.5 mM EDTA, 0.02 % NaN3, pH: 6.8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Feb 22
|
Domain deletion constructs of hHR23B
Sarasi Galagedera
|
| RgGuinier |
4.1 |
nm |
| Dmax |
17.5 |
nm |
| VolumePorod |
79 |
nm3 |
|
|
UniProt ID: P54727 (1-409) UV excision repair protein RAD23 homolog B (∆274-306)
|
|
|
|
| Sample: |
UV excision repair protein RAD23 homolog B (∆274-306) monomer, 39 kDa Homo sapiens protein
|
| Buffer: |
20 mM sodium phosphate, 0.5 mM EDTA, 0.02 % NaN3, pH: 6.8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Feb 22
|
Domain deletion constructs of hHR23B
Sarasi Galagedera
|
| RgGuinier |
4.6 |
nm |
| Dmax |
24.2 |
nm |
| VolumePorod |
103 |
nm3 |
|
|
UniProt ID: P54727 (1-409) UV excision repair protein RAD23 homolog B (∆186-231, ∆362-409)
|
|
|
|
| Sample: |
UV excision repair protein RAD23 homolog B (∆186-231, ∆362-409) monomer, 33 kDa Homo sapiens protein
|
| Buffer: |
20 mM sodium phosphate, 0.5 mM EDTA, 0.02 % NaN3, pH: 6.8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Feb 22
|
Domain deletion constructs of hHR23B
Sarasi Galagedera
|
| RgGuinier |
4.9 |
nm |
| Dmax |
25.5 |
nm |
| VolumePorod |
100 |
nm3 |
|
|
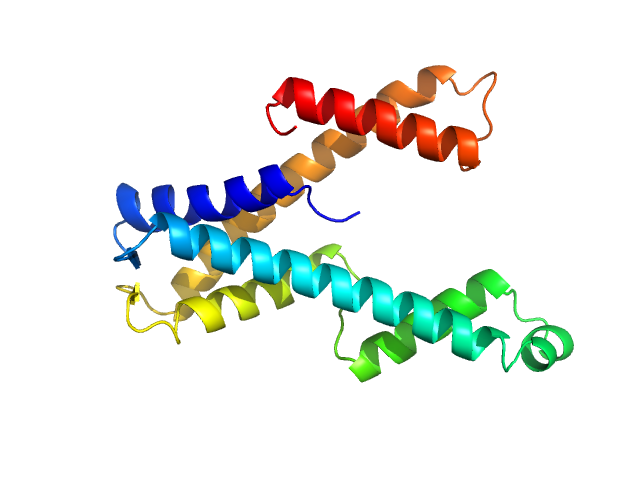
UniProt ID: O67633 (1-183) DUF507 family protein
|
|
|
|
| Sample: |
DUF507 family protein monomer, 22 kDa Aquifex aeolicus (strain … protein
|
| Buffer: |
20 mM Hepes pH 7.2, 400 mM NaCl, pH: 7.2 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2021 Dec 7
|
Crystal structure of domain of unknown function 507 (DUF507) reveals a new protein fold.
Sci Rep 13(1):13496 (2023)
McKay CE, Cheng J, Tanner JJ
|
| RgGuinier |
2.3 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
39 |
nm3 |
|
|
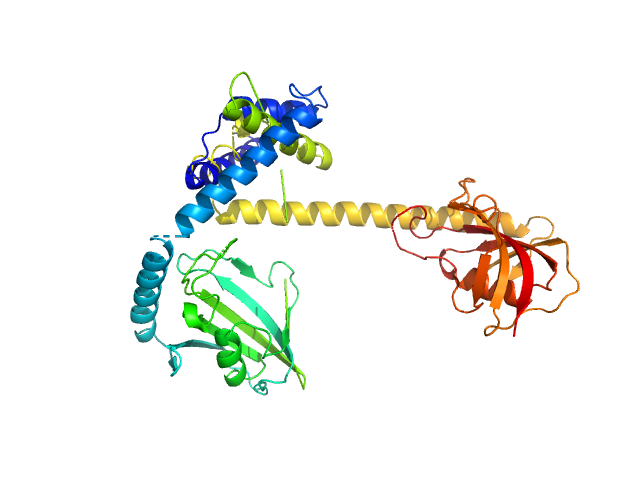
UniProt ID: Q5ZXE0 (21-233) Outer membrane protein MIP
|
|
|
|
| Sample: |
Outer membrane protein MIP dimer, 46 kDa Legionella pneumophila subsp. … protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, 10 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Mar 16
|
Legionella pneumophila macrophage infectivity potentiator protein appendage domains modulate protein dynamics and inhibitor binding
International Journal of Biological Macromolecules :126366 (2023)
Wiedemann C, Whittaker J, Carrillo V, Goretzki B, Dajka M, Tebbe F, Harder J, Krajczy P, Joseph B, Hausch F, Guskov A, Hellmich U
|
| RgGuinier |
3.1 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
66 |
nm3 |
|
|