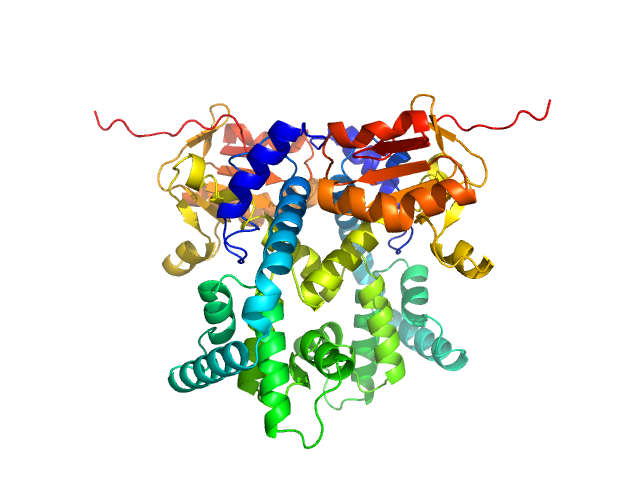
UniProt ID: Q9Y2V0 (1-281) CDAN1-interacting nuclease 1
|
|
|
|
| Sample: |
CDAN1-interacting nuclease 1 dimer, 67 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris HCl, 150 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpoint 2.0, Institute of Biotechnology, Czech Academy of Sciences/Centre of Molecular Structure on 2021 Sep 2
|
Structural insights into inherited anemia CDA-I: disease-associated mutations disrupt Codanin1-CDIN1 complex
Tomas Brom
|
| RgGuinier |
3.0 |
nm |
| Dmax |
8.2 |
nm |
| VolumePorod |
84 |
nm3 |
|
|
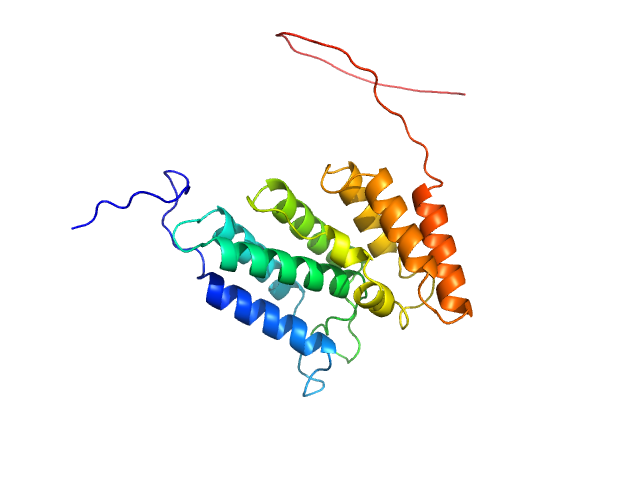
UniProt ID: Q8IWY9 (1005-1227) Codanin-1
|
|
|
|
| Sample: |
Codanin-1 monomer, 25 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris HCl, 150 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpoint 2.0, Institute of Biotechnology, Czech Academy of Sciences/Centre of Molecular Structure on 2022 Mar 5
|
Structural insights into inherited anemia CDA-I: disease-associated mutations disrupt Codanin1-CDIN1 complex
Tomas Brom
|
| RgGuinier |
2.7 |
nm |
| Dmax |
7.5 |
nm |
| VolumePorod |
54 |
nm3 |
|
|
UniProt ID: Q8IWY9 (1005-1227) Codanin-1
|
|
|

|
| Sample: |
Codanin-1 monomer, 25 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris HCl, 150 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpoint 2.0, Institute of Biotechnology, Czech Academy of Sciences/Centre of Molecular Structure on 2022 Mar 5
|
Structural insights into inherited anemia CDA-I: disease-associated mutations disrupt Codanin1-CDIN1 complex
Tomas Brom
|
| RgGuinier |
4.1 |
nm |
| Dmax |
10.7 |
nm |
| VolumePorod |
220 |
nm3 |
|
|
UniProt ID: O75031 (2-334) Heat shock factor 2-binding protein
|
|
|
|
| Sample: |
Heat shock factor 2-binding protein tetramer, 150 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris-HCl pH 7.5, 250 mM NaCl, and 5 mM β-mercaptoethanol, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2020 Oct 2
|
BRCA2-HSF2BP oligomeric ring disassembly by BRME1 promotes homologous recombination.
Sci Adv 9(43):eadi7352 (2023)
Ghouil R, Miron S, Sato K, Ristic D, van Rossum-Fikkert SE, Legrand P, Ouldali M, Winter JM, Ropars V, David G, Arteni AA, Wyman C, Knipscheer P, Kanaar R, Zelensky AN, Zinn-Justin S
|
| RgGuinier |
8.5 |
nm |
| Dmax |
25.0 |
nm |
| VolumePorod |
364 |
nm3 |
|
|
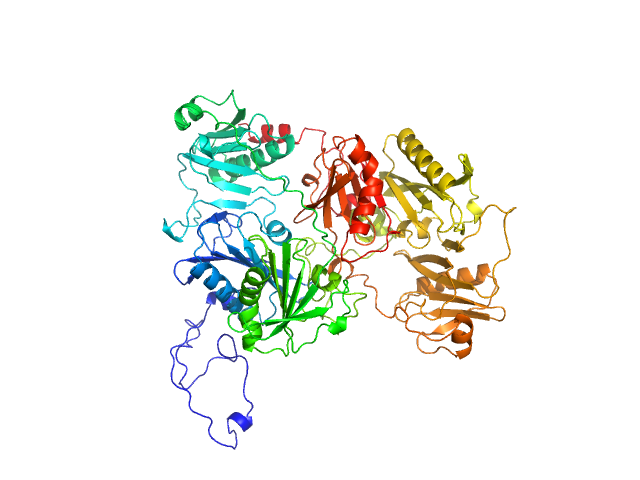
UniProt ID: P06396 (28-782) Gelsolin
|
|
|
|
| Sample: |
Gelsolin monomer, 85 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, 1 mM EDTA, pH: 7.4 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2020 Feb 24
|
Accurate and Efficient SAXS/SANS Implementation Including Solvation Layer Effects Suitable for Molecular Simulations.
J Chem Theory Comput (2023)
Ballabio F, Paissoni C, Bollati M, de Rosa M, Capelli R, Camilloni C
|
| RgGuinier |
3.2 |
nm |
| Dmax |
11.1 |
nm |
| VolumePorod |
128 |
nm3 |
|
|
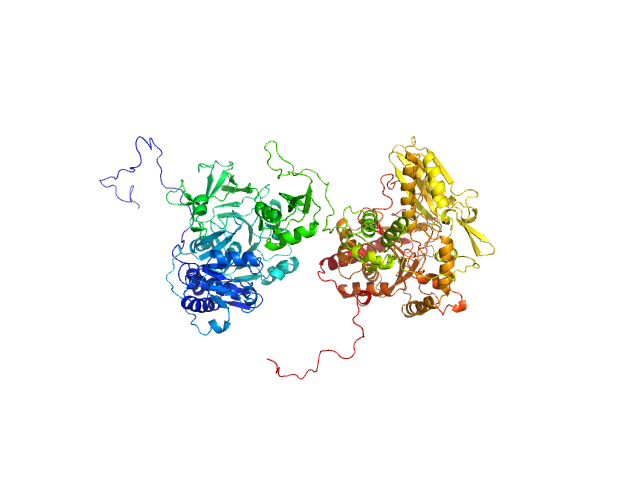
UniProt ID: P0C061 (1-1098) Gramicidin S synthase 1 (W239S)
|
|
|
|
| Sample: |
Gramicidin S synthase 1 (W239S) monomer, 127 kDa Aneurinibacillus migulanus protein
|
| Buffer: |
20 mM Tris pH 7.5, 100 mM NaCl, 10 mM MgCl2, 2% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2023 Mar 14
|
Resting state of GrsA is a combination of two constitutively inactive isoforms that manifest long-range allosteric effects in presence of aminoacid and ATP
Raktim Roy
|
| RgGuinier |
4.1 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
187 |
nm3 |
|
|
UniProt ID: P0C062 (1-1098) Gramicidin S synthase 1 (W239S)
|
|
|
|
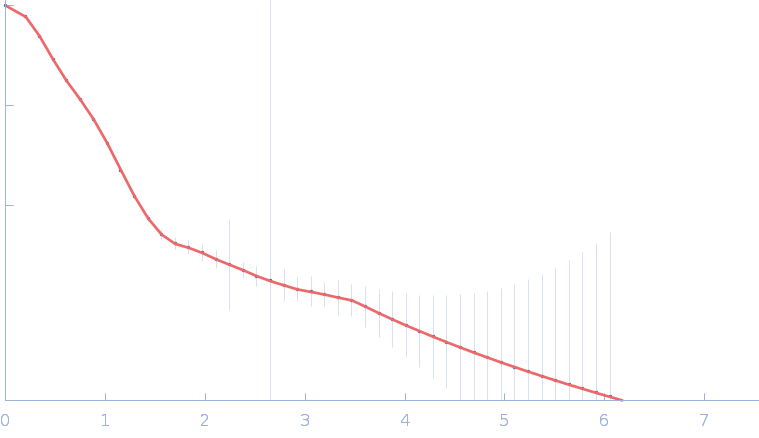
| Sample: |
Gramicidin S synthase 1 (W239S) monomer, 126 kDa Brevibacillus brevis protein
|
| Buffer: |
20 mM Tris pH 7.5, 100 mM NaCl, 10 mM MgCl2, 2% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2023 Mar 14
|
Resting state of GrsA is a combination of two constitutively inactive isoforms that manifest long-range allosteric effects in presence of aminoacid and ATP
Raktim Roy
|
| RgGuinier |
4.3 |
nm |
| Dmax |
15.4 |
nm |
| VolumePorod |
186 |
nm3 |
|
|
UniProt ID: P0C062 (1-1098) Gramicidin S synthase 1 (W239S)
|
|
|
|
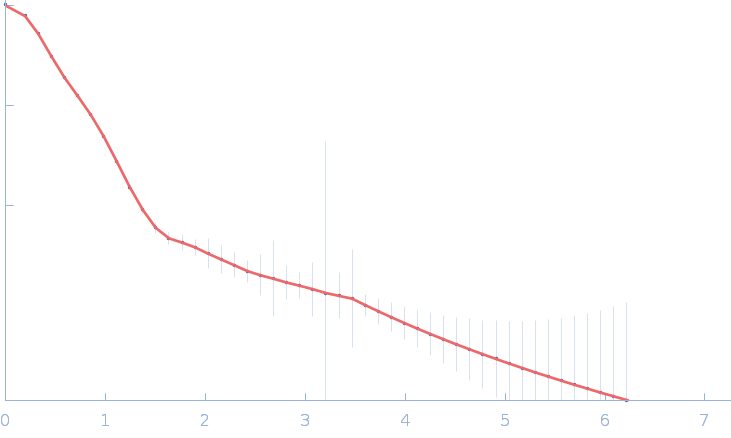
| Sample: |
Gramicidin S synthase 1 (W239S) monomer, 126 kDa Brevibacillus brevis protein
|
| Buffer: |
20 mM Tris pH 7.5, 100 mM NaCl, 10 mM MgCl2, 2% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2023 Mar 14
|
Resting state of GrsA is a combination of two constitutively inactive isoforms that manifest long-range allosteric effects in presence of aminoacid and ATP
Raktim Roy
|
| RgGuinier |
4.2 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
186 |
nm3 |
|
|
UniProt ID: P0C062 (1-1098) Gramicidin S synthase 1 (W239S)
|
|
|
|
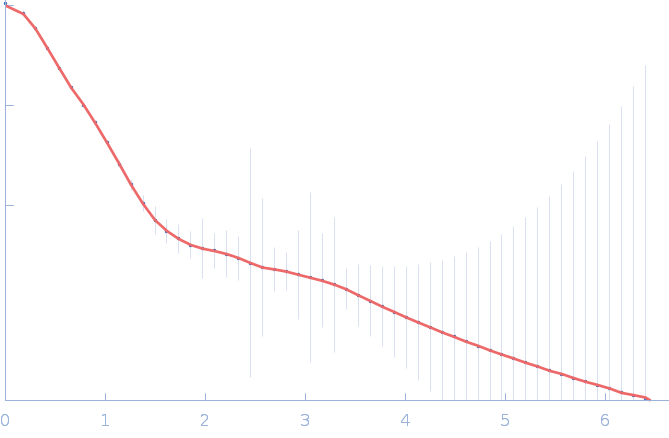
| Sample: |
Gramicidin S synthase 1 (W239S) monomer, 126 kDa Brevibacillus brevis protein
|
| Buffer: |
20 mM Tris pH 7.5, 100 mM NaCl, 10 mM MgCl2, 2% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2023 Mar 14
|
Resting state of GrsA is a combination of two constitutively inactive isoforms that manifest long-range allosteric effects in presence of aminoacid and ATP
Raktim Roy
|
| RgGuinier |
4.1 |
nm |
| Dmax |
17.5 |
nm |
| VolumePorod |
181 |
nm3 |
|
|
UniProt ID: P0C062 (1-1098) Gramicidin S synthase 1 (W239S)
|
|
|
|
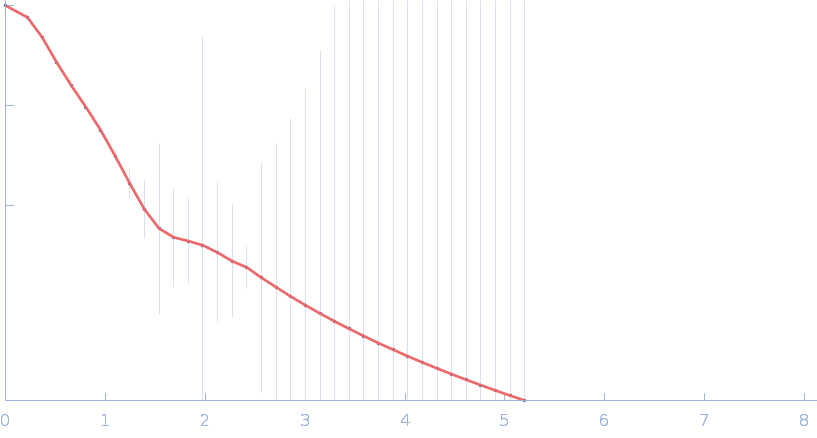
| Sample: |
Gramicidin S synthase 1 (W239S) monomer, 126 kDa Brevibacillus brevis protein
|
| Buffer: |
20 mM Tris pH 7.5, 100 mM NaCl, 10 mM MgCl2, 2% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2023 Mar 14
|
Resting state of GrsA is a combination of two constitutively inactive isoforms that manifest long-range allosteric effects in presence of aminoacid and ATP
Raktim Roy
|
| RgGuinier |
4.0 |
nm |
| Dmax |
14.3 |
nm |
| VolumePorod |
175 |
nm3 |
|
|