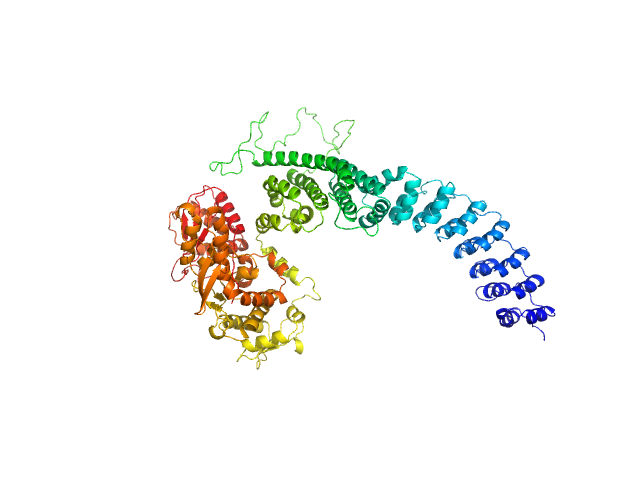
UniProt ID: Q8IYU2 (22-909) E3 ubiquitin-protein ligase HACE1
|
|
|
|
| Sample: |
E3 ubiquitin-protein ligase HACE1 monomer, 100 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 50 mM NaCl, 5 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2023 Apr 21
|
Structural mechanisms of autoinhibition and substrate recognition by the ubiquitin ligase HACE1
Nature Structural & Molecular Biology (2024)
Düring J, Wolter M, Toplak J, Torres C, Dybkov O, Fokkens T, Bohnsack K, Urlaub H, Steinchen W, Dienemann C, Lorenz S
|
| RgGuinier |
4.6 |
nm |
| Dmax |
17.8 |
nm |
| VolumePorod |
219 |
nm3 |
|
|
UniProt ID: Q4K5N8 (1-183) Heme acquisition protein HasAp
|
|
|

|
| Sample: |
Heme acquisition protein HasAp dimer, 38 kDa Pseudomonas fluorescens (strain … protein
|
| Buffer: |
50 mM CHES, 5 % glycerol, pH: 9.5 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2023 Feb 24
|
Heme-substituted protein assembly bridged by synthetic porphyrin: achieving controlled configuration while maintaining rotational freedom
RSC Advances 14(13):8829-8836 (2024)
Inaba H, Shisaka Y, Ariyasu S, Sakakibara E, Ueda G, Aiba Y, Shimizu N, Sugimoto H, Shoji O
|
| RgGuinier |
2.8 |
nm |
| Dmax |
9.1 |
nm |
| VolumePorod |
54 |
nm3 |
|
|
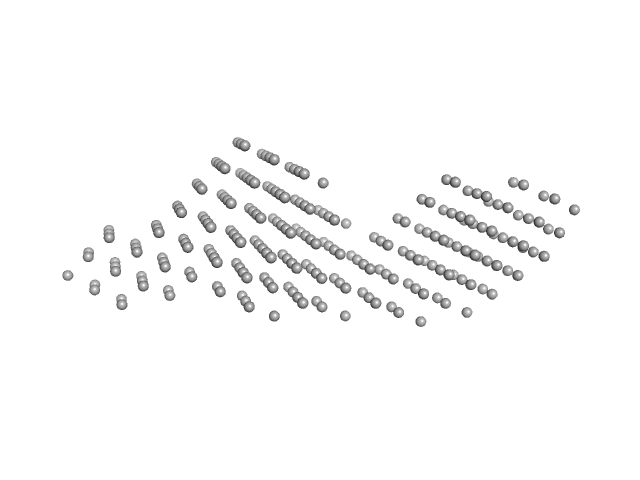
UniProt ID: P18035 (1-181) Protein TrbB (GST-fusion)
|
|
|
|
| Sample: |
Protein TrbB (GST-fusion) dimer, 93 kDa Escherichia coli (strain … protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, 5% glycerol, 0.05% NP40, pH: 7 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Feb 23
|
Structural insights into the disulfide isomerase and chaperone activity of TrbB of the F plasmid type IV secretion system
Current Research in Structural Biology 8:100156 (2024)
Apostol A, Bragagnolo N, Rodriguez C, Audette G
|
| RgGuinier |
4.0 |
nm |
| Dmax |
18.0 |
nm |
| VolumePorod |
153 |
nm3 |
|
|
UniProt ID: A0A1Z4BZH1 (1-81) NHLP leader peptide family natural product (I56V)
|
|
|
|
| Sample: |
NHLP leader peptide family natural product (I56V) monomer, 9 kDa Methylovulum psychrotolerans protein
|
| Buffer: |
20 mM sodium phosphate (pH 7.5), 100 mM NaCl and 48 µM FAD, pH: 7.5 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-2000, Pennsylvania State University on 2023 Jul 28
|
Disordered regions in proteusin peptides guide post-translational modification by a flavin-dependent RiPP brominase.
Nat Commun 15(1):1265 (2024)
Nguyen NA, Vidya FNU, Yennawar NH, Wu H, McShan AC, Agarwal V
|
| RgGuinier |
1.9 |
nm |
| Dmax |
5.5 |
nm |
| VolumePorod |
39 |
nm3 |
|
|
UniProt ID: Q8TDB6 (101-200) E3 ubiquitin-protein ligase DTX3L
|
|
|
|
| Sample: |
E3 ubiquitin-protein ligase DTX3L tetramer, 46 kDa Homo sapiens protein
|
| Buffer: |
30 mM HEPES, 350 mM NaCl, 5% (v/v) glycerol, 0.5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2021 Dec 6
|
Oligomerisation mediated by the D2 domain of DTX3L is critical for DTX3L-PARP9 reading function of mono-ADP-ribosylated androgen receptor.
bioRxiv (2023)
Vela-Rodríguez C, Yang C, Alanen HI, Eki R, Abbas TA, Maksimainen MM, Glumoff T, Duman R, Wagner A, Paschal BM, Lehtiö L
|
| RgGuinier |
2.9 |
nm |
| Dmax |
112.0 |
nm |
| VolumePorod |
91 |
nm3 |
|
|
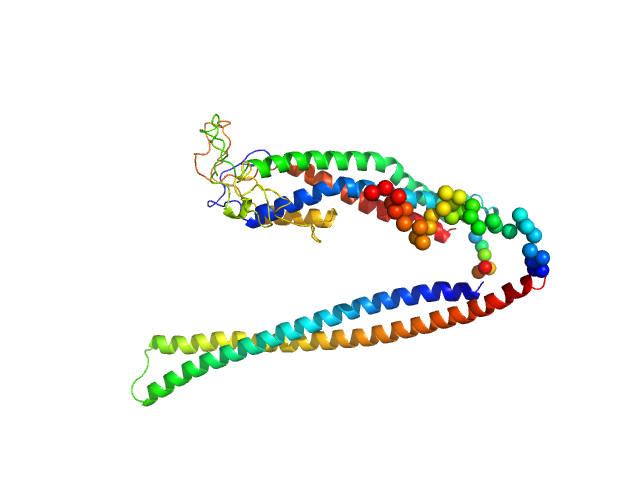
UniProt ID: Q26769 (29-468) Invariant surface glycoprotein (N134A)
|
|
|
|
| Sample: |
Invariant surface glycoprotein (N134A) monomer, 49 kDa Trypanosoma brucei protein
|
| Buffer: |
20 mM Tris-HCl, 75 mM KCl, pH: 7.6 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Aug 18
|
Trypanosoma brucei Invariant Surface Glycoprotein 75 Is an Immunoglobulin Fc Receptor Inhibiting Complement Activation and Antibody-Mediated Cellular Phagocytosis.
J Immunol (2024)
Mikkelsen JH, Stødkilde K, Jensen MP, Hansen AG, Wu Q, Lorentzen J, Graversen JH, Andersen GR, Fenton RA, Etzerodt A, Thiel S, Andersen CBF
|
| RgGuinier |
3.6 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
127 |
nm3 |
|
|
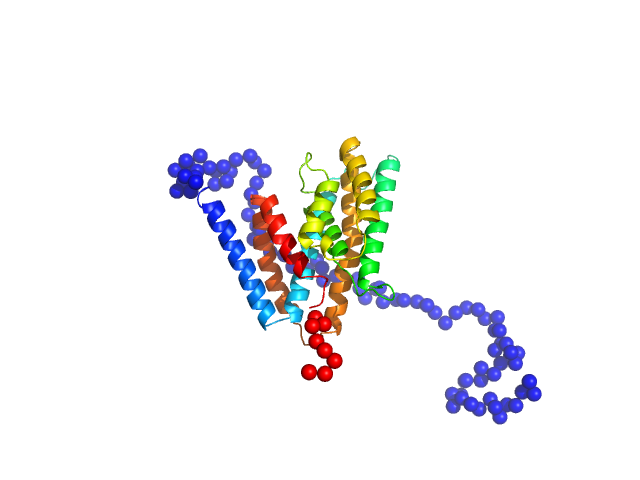
UniProt ID: Q9Z7M7 (67-382) Uncharacterized protein
|
|
|
|
| Sample: |
Uncharacterized protein monomer, 35 kDa Chlamydia pneumoniae protein
|
| Buffer: |
137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, 3 %Glycerol, pH: 8.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Dec 14
|
The Chlamydia pneumoniae effector SemD exploits its host’s endocytic machinery by structural and functional mimicry
Nature Communications 15(1) (2024)
Kocher F, Applegate V, Reiners J, Port A, Spona D, Hänsch S, Mirzaiebadizi A, Ahmadian M, Smits S, Hegemann J, Mölleken K
|
| RgGuinier |
2.8 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
67 |
nm3 |
|
|
UniProt ID: O08816 (141-273) Actin nucleation-promoting factor WASL
|
|
|
|
| Sample: |
Actin nucleation-promoting factor WASL monomer, 19 kDa Rattus norvegicus protein
|
| Buffer: |
137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, 3 %Glycerol, pH: 8.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Dec 14
|
The Chlamydia pneumoniae effector SemD exploits its host’s endocytic machinery by structural and functional mimicry
Nature Communications 15(1) (2024)
Kocher F, Applegate V, Reiners J, Port A, Spona D, Hänsch S, Mirzaiebadizi A, Ahmadian M, Smits S, Hegemann J, Mölleken K
|
| RgGuinier |
3.0 |
nm |
| Dmax |
9.9 |
nm |
| VolumePorod |
19 |
nm3 |
|
|
UniProt ID: Q9Y5X1 (1-160) Sorting nexin-9
|
|
|
|
| Sample: |
Sorting nexin-9 monomer, 19 kDa Homo sapiens protein
|
| Buffer: |
137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, 3 %Glycerol, pH: 8.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2023 Mar 20
|
The Chlamydia pneumoniae effector SemD exploits its host’s endocytic machinery by structural and functional mimicry
Nature Communications 15(1) (2024)
Kocher F, Applegate V, Reiners J, Port A, Spona D, Hänsch S, Mirzaiebadizi A, Ahmadian M, Smits S, Hegemann J, Mölleken K
|
| RgGuinier |
2.2 |
nm |
| Dmax |
7.1 |
nm |
| VolumePorod |
42 |
nm3 |
|
|
UniProt ID: Q9Z7M7 (67-382) Uncharacterized protein
UniProt ID: Q9Y5X1 (1-160) Sorting nexin-9
|
|
|
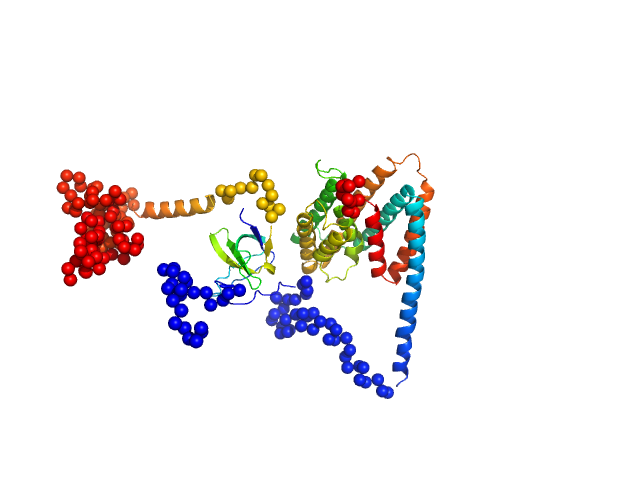
|
| Sample: |
Uncharacterized protein monomer, 35 kDa Chlamydia pneumoniae protein
Sorting nexin-9 monomer, 19 kDa Homo sapiens protein
|
| Buffer: |
137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, 3 %Glycerol, pH: 8.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2023 Mar 20
|
The Chlamydia pneumoniae effector SemD exploits its host’s endocytic machinery by structural and functional mimicry
Nature Communications 15(1) (2024)
Kocher F, Applegate V, Reiners J, Port A, Spona D, Hänsch S, Mirzaiebadizi A, Ahmadian M, Smits S, Hegemann J, Mölleken K
|
| RgGuinier |
3.4 |
nm |
| Dmax |
12.2 |
nm |
| VolumePorod |
94 |
nm3 |
|
|