UniProt ID: Q2FHL2 (1-548) Uncharacterized protein SAUSA300_1119
|
|
|

|
| Sample: |
Uncharacterized protein SAUSA300_1119 dimer, 125 kDa Staphylococcus aureus (strain … protein
|
| Buffer: |
50 mM Tris, 150 mM KCl, 1 mM TCEP, 5% glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Oct 5
|
Molecular insights into the structure and function of the Staphylococcus aureus fatty acid kinase.
J Biol Chem 300(12):107920 (2024)
Myers MJ, Xu Z, Ryan BJ, DeMars ZR, Ridder MJ, Johnson DK, Krute CN, Flynn TS, Kashipathy MM, Battaile KP, Schnicker N, Lovell S, Freudenthal BD, Bose JL
|
| RgGuinier |
4.3 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
219 |
nm3 |
|
|
UniProt ID: P0DN69 (1-110) Glycine cleavage system H-like protein
|
|
|
|
| Sample: |
Glycine cleavage system H-like protein monomer, 13 kDa Streptococcus pyogenes serotype … protein
|
| Buffer: |
50 mM Tris-HCl, 200 mM NaCl, 1 mM DTT, 5% (v/v) glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2016 Jul 20
|
Evolutionary and molecular basis of ADP-ribosylation reversal by zinc-dependent macrodomains
Journal of Biological Chemistry :107770 (2024)
Ariza A, Liu Q, Cowieson N, Ahel I, Filippov D, Rack J
|
| RgGuinier |
1.5 |
nm |
| Dmax |
5.0 |
nm |
| VolumePorod |
22 |
nm3 |
|
|
UniProt ID: P0DN70 (1-270) Protein-ADP-ribose hydrolase (D13G, Y23S, T61A, I114S, R177H, I246T)
|
|
|
|
| Sample: |
Protein-ADP-ribose hydrolase (D13G, Y23S, T61A, I114S, R177H, I246T) monomer, 30 kDa Streptococcus pyogenes serotype … protein
|
| Buffer: |
50 mM Tris-HCl, 200 mM NaCl, 1 mM DTT, 5% (v/v) glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2016 Jul 20
|
Evolutionary and molecular basis of ADP-ribosylation reversal by zinc-dependent macrodomains
Journal of Biological Chemistry :107770 (2024)
Ariza A, Liu Q, Cowieson N, Ahel I, Filippov D, Rack J
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.3 |
nm |
| VolumePorod |
34 |
nm3 |
|
|
UniProt ID: P0DN69 (1-110) Glycine cleavage system H-like protein
UniProt ID: P0DN70 (1-270) Protein-ADP-ribose hydrolase (D13G, Y23S, T61A, I114S, R177H, I246T)
|
|
|
|
| Sample: |
Glycine cleavage system H-like protein monomer, 13 kDa Streptococcus pyogenes serotype … protein
Protein-ADP-ribose hydrolase (D13G, Y23S, T61A, I114S, R177H, I246T) monomer, 30 kDa Streptococcus pyogenes serotype … protein
|
| Buffer: |
50 mM Tris-HCl, 200 mM NaCl, 1 mM DTT, 5% (v/v) glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2016 Jul 20
|
Evolutionary and molecular basis of ADP-ribosylation reversal by zinc-dependent macrodomains
Journal of Biological Chemistry :107770 (2024)
Ariza A, Liu Q, Cowieson N, Ahel I, Filippov D, Rack J
|
| RgGuinier |
2.4 |
nm |
| Dmax |
8.4 |
nm |
| VolumePorod |
53 |
nm3 |
|
|
UniProt ID: None (None-None) Cereblon-midi
|
|
|
|
| Sample: |
Cereblon-midi monomer, 37 kDa protein
|
| Buffer: |
20 mM Hepes, 500 mM NaCl, 0.5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 Mar 1
|
Design of a Cereblon construct for crystallographic and biophysical studies of protein degraders
Nature Communications 15(1) (2024)
Kroupova A, Spiteri V, Rutter Z, Furihata H, Darren D, Ramachandran S, Chakraborti S, Haubrich K, Pethe J, Gonzales D, Wijaya A, Rodriguez-Rios M, Sturbaut M, Lynch D, Farnaby W, Nakasone M, Zollman D, Ciulli A
|
| RgGuinier |
2.7 |
nm |
| Dmax |
8.6 |
nm |
| VolumePorod |
61 |
nm3 |
|
|
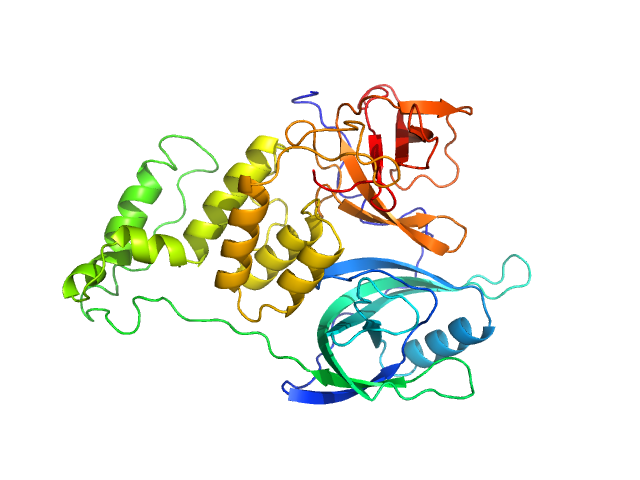
UniProt ID: None (None-None) Cereblon-midi
UniProt ID: None (None-None) mezigdomide
|
|
|
|
| Sample: |
Cereblon-midi monomer, 37 kDa protein
Mezigdomide monomer, 1 kDa synthetic construct
|
| Buffer: |
20 mM Hepes, 500 mM NaCl, 0.5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 Mar 1
|
Design of a Cereblon construct for crystallographic and biophysical studies of protein degraders
Nature Communications 15(1) (2024)
Kroupova A, Spiteri V, Rutter Z, Furihata H, Darren D, Ramachandran S, Chakraborti S, Haubrich K, Pethe J, Gonzales D, Wijaya A, Rodriguez-Rios M, Sturbaut M, Lynch D, Farnaby W, Nakasone M, Zollman D, Ciulli A
|
| RgGuinier |
2.3 |
nm |
| Dmax |
7.3 |
nm |
| VolumePorod |
60 |
nm3 |
|
|
UniProt ID: None (None-None) Cereblon-midi
UniProt ID: None (None-None) pomalidomide
|
|
|
|
| Sample: |
Cereblon-midi monomer, 37 kDa protein
Pomalidomide monomer, 0 kDa synthetic construct
|
| Buffer: |
20 mM Hepes, 500 mM NaCl, 0.5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 Mar 1
|
Design of a Cereblon construct for crystallographic and biophysical studies of protein degraders
Nature Communications 15(1) (2024)
Kroupova A, Spiteri V, Rutter Z, Furihata H, Darren D, Ramachandran S, Chakraborti S, Haubrich K, Pethe J, Gonzales D, Wijaya A, Rodriguez-Rios M, Sturbaut M, Lynch D, Farnaby W, Nakasone M, Zollman D, Ciulli A
|
| RgGuinier |
2.3 |
nm |
| Dmax |
6.9 |
nm |
| VolumePorod |
63 |
nm3 |
|
|
UniProt ID: None (None-None) Cereblon-midi
UniProt ID: None (None-None) Iberdomide
|
|
|
|
| Sample: |
Cereblon-midi monomer, 37 kDa protein
Iberdomide monomer, 0 kDa synthetic construct
|
| Buffer: |
20 mM Hepes, 500 mM NaCl, 0.5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 Mar 1
|
Design of a Cereblon construct for crystallographic and biophysical studies of protein degraders
Nature Communications 15(1) (2024)
Kroupova A, Spiteri V, Rutter Z, Furihata H, Darren D, Ramachandran S, Chakraborti S, Haubrich K, Pethe J, Gonzales D, Wijaya A, Rodriguez-Rios M, Sturbaut M, Lynch D, Farnaby W, Nakasone M, Zollman D, Ciulli A
|
| RgGuinier |
2.4 |
nm |
| Dmax |
7.7 |
nm |
| VolumePorod |
68 |
nm3 |
|
|
UniProt ID: None (None-None) Cereblon-midi
UniProt ID: None (None-None) Lenalidomide
|
|
|
|
| Sample: |
Cereblon-midi monomer, 37 kDa protein
Lenalidomide monomer, 0 kDa synthetic construct
|
| Buffer: |
20 mM Hepes, 500 mM NaCl, 0.5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 Mar 1
|
Design of a Cereblon construct for crystallographic and biophysical studies of protein degraders
Nature Communications 15(1) (2024)
Kroupova A, Spiteri V, Rutter Z, Furihata H, Darren D, Ramachandran S, Chakraborti S, Haubrich K, Pethe J, Gonzales D, Wijaya A, Rodriguez-Rios M, Sturbaut M, Lynch D, Farnaby W, Nakasone M, Zollman D, Ciulli A
|
| RgGuinier |
2.2 |
nm |
| Dmax |
6.2 |
nm |
| VolumePorod |
59 |
nm3 |
|
|
UniProt ID: Q9H171 (1-165) Z-DNA-binding protein 1
|
|
|

|
| Sample: |
Z-DNA-binding protein 1 monomer, 18 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 100 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2018 Mar 23
|
ZBP1 condensate formation synergizes Z-NAs recognition and signal transduction.
Cell Death Dis 15(7):487 (2024)
Xie F, Wu D, Huang J, Liu X, Shen Y, Huang J, Su Z, Li J
|
| RgGuinier |
2.1 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
19 |
nm3 |
|
|