UniProt ID: E7BLH6 (387-536) HbP1 (K526A)
|
|
|
|
| Sample: |
HbP1 (K526A) trimer, 56 kDa Legionella pneumophila protein
|
| Buffer: |
20 mM Tris–HCl pH 8.0, 200 mM NaCl, 5 mM EDTA, pH: |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Jul 29
|
The Legionella collagen-like protein employs a distinct binding mechanism for the recognition of host glycosaminoglycans.
Nat Commun 15(1):4912 (2024)
Rehman S, Antonovic AK, McIntire IE, Zheng H, Cleaver L, Baczynska M, Adams CO, Portlock T, Richardson K, Shaw R, Oregioni A, Mastroianni G, Whittaker SB, Kelly G, Lorenz CD, Fornili A, Cianciotto NP, Garnett JA
|
| RgGuinier |
2.8 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
104 |
nm3 |
|
|
UniProt ID: P61825 (844-976) Structural polyprotein (Capsid protein VP3: K947R; Δ756-843; Δ977-1012)
|
|
|
|
| Sample: |
Structural polyprotein (Capsid protein VP3: K947R; Δ756-843; Δ977-1012) monomer, 18 kDa Infectious bursal disease … protein
|
| Buffer: |
50 mM TRIS, 500 mM NaCl, 2 mM DTT,, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Jul 2
|
Infectious Bursal Disease Virus VP3
Diego Ferrero
|
| RgGuinier |
2.5 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
21 |
nm3 |
|
|
UniProt ID: P61825 (756-976) Structural polyprotein (Capsid protein VP3: K947R; Δ977-1012)
|
|
|
|
| Sample: |
Structural polyprotein (Capsid protein VP3: K947R; Δ977-1012) dimer, 55 kDa Infectious bursal disease … protein
|
| Buffer: |
50 mM TRIS, 500 mM NaCl, 2 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Jul 1
|
Infectious Bursal Disease Virus VP3
Diego Ferrero
|
| RgGuinier |
4.1 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
110 |
nm3 |
|
|
UniProt ID: P43405 (6-269) Tyrosine-protein kinase SYK
|
|
|
|
| Sample: |
Tyrosine-protein kinase SYK monomer, 30 kDa Homo sapiens protein
|
| Buffer: |
10 mM HEPES, 150 mM NaCl, 1 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 Nov 13
|
The mechanism of allosteric activation of SYK kinase derived from multiple phospho-ITAM-bound structures
Structure (2024)
Bradshaw W, Harris G, Gileadi O, Katis V
|
| RgGuinier |
2.2 |
nm |
| Dmax |
6.9 |
nm |
| VolumePorod |
49 |
nm3 |
|
|
UniProt ID: P43405 (6-269) Tyrosine-protein kinase SYK
UniProt ID: P30273 (62-81) High affinity immunoglobulin epsilon receptor subunit gamma
|
|
|
|
| Sample: |
Tyrosine-protein kinase SYK monomer, 30 kDa Homo sapiens protein
High affinity immunoglobulin epsilon receptor subunit gamma monomer, 2 kDa Homo sapiens protein
|
| Buffer: |
10 mM HEPES, 150 mM NaCl, 1 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 Nov 13
|
The mechanism of allosteric activation of SYK kinase derived from multiple phospho-ITAM-bound structures
Structure (2024)
Bradshaw W, Harris G, Gileadi O, Katis V
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.0 |
nm |
| VolumePorod |
54 |
nm3 |
|
|
UniProt ID: P43405 (6-269) Tyrosine-protein kinase SYK
UniProt ID: P09693 (157-176) T-cell surface glycoprotein CD3 gamma chain
|
|
|
|
| Sample: |
Tyrosine-protein kinase SYK monomer, 30 kDa Homo sapiens protein
T-cell surface glycoprotein CD3 gamma chain monomer, 3 kDa Homo sapiens protein
|
| Buffer: |
10 mM HEPES, 150 mM NaCl, 1 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 Nov 13
|
The mechanism of allosteric activation of SYK kinase derived from multiple phospho-ITAM-bound structures
Structure (2024)
Bradshaw W, Harris G, Gileadi O, Katis V
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.1 |
nm |
| VolumePorod |
56 |
nm3 |
|
|
UniProt ID: P43405 (6-269) Tyrosine-protein kinase SYK
UniProt ID: O43914 (88-107) TYRO protein tyrosine kinase-binding protein
|
|
|
|
| Sample: |
Tyrosine-protein kinase SYK monomer, 30 kDa Homo sapiens protein
TYRO protein tyrosine kinase-binding protein monomer, 2 kDa Homo sapiens protein
|
| Buffer: |
10 mM HEPES, 150 mM NaCl, 1 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 Nov 13
|
The mechanism of allosteric activation of SYK kinase derived from multiple phospho-ITAM-bound structures
Structure (2024)
Bradshaw W, Harris G, Gileadi O, Katis V
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.1 |
nm |
| VolumePorod |
53 |
nm3 |
|
|
UniProt ID: Q9P8F4 (1-860) beta-glucosidase
|
|
|

|
| Sample: |
Beta-glucosidase dimer, 187 kDa Aspergillus niger protein
|
| Buffer: |
50 mM sodium acetate, pH: 5 |
| Experiment: |
SAXS
data collected at 16-ID (LiX), National Synchrotron Light Source II (NSLS-II) on 2024 Feb 15
|
Aspergillus niger beta-glucosidase (bgl1)
Estella Yee
|
| RgGuinier |
4.2 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
253 |
nm3 |
|
|
UniProt ID: Q8WPR6 (480-859) adenylate cyclase
|
|
|
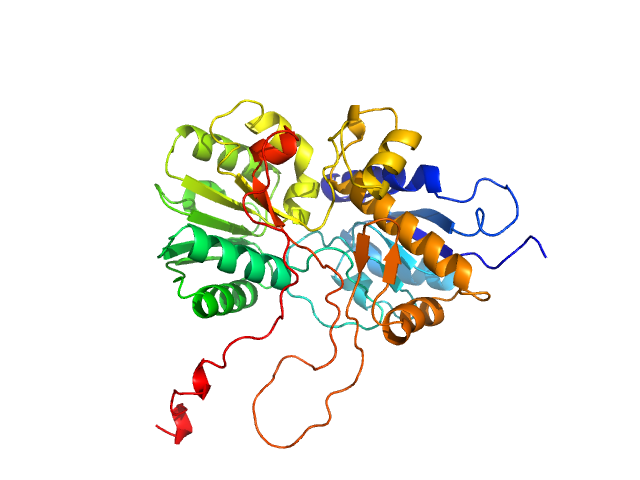
|
| Sample: |
Adenylate cyclase monomer, 43 kDa Trypanosoma brucei protein
|
| Buffer: |
50 mM Tris-HCl, 500 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2023 Sep 23
|
Biophysical analysis of the membrane-proximal Venus Flytrap domain of ESAG4 receptor-like adenylate cyclase from Trypanosoma brucei.
Mol Biochem Parasitol 260:111653 (2024)
Alves DO, Geens R, da Silva Arruda HR, Jennen L, Corthaut S, Wuyts E, de Andrade GC, Prosdocimi F, Cordeiro Y, Pires JR, Vieira LR, de Oliveira GAP, Sterckx YG, Salmon D
|
| RgGuinier |
2.3 |
nm |
| Dmax |
7.6 |
nm |
| VolumePorod |
76 |
nm3 |
|
|
UniProt ID: P0AEX9 (27-384) Maltose/maltodextrin-binding periplasmic protein (D108A, K109A, E198A, N199A, K265A)
UniProt ID: P9WPH5 (19-409) Uncharacterized protein Rv2242
|
|
|
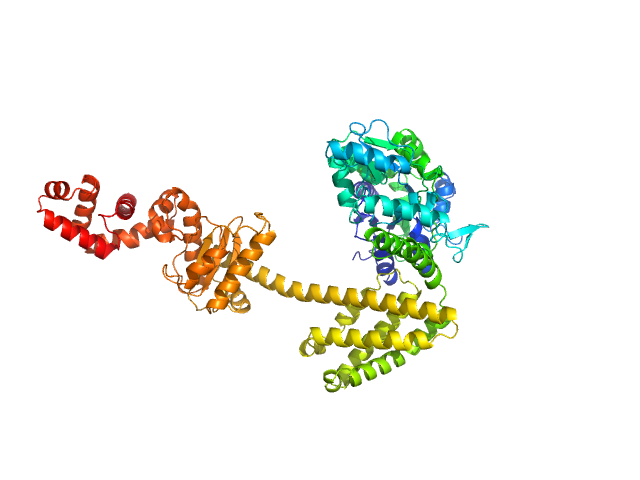
|
| Sample: |
Maltose/maltodextrin-binding periplasmic protein (D108A, K109A, E198A, N199A, K265A) monomer, 40 kDa Escherichia coli (strain … protein
Uncharacterized protein Rv2242 monomer, 43 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
50 mM Tris pH 7.5, 150 mM NaCl, pH: |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2024 Apr 3
|
Domain architecture of the Mycobacterium tuberculosis MabR (Rv2242), a member of the PucR transcription factor family
Heliyon :e40494 (2024)
Megalizzi V, Tanina A, Grosse C, Mirgaux M, Legrand P, Mirandela G, Wohlkönig A, Bifani P, Wintjens R
|
| RgGuinier |
2.8 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
113 |
nm3 |
|
|