UniProt ID: O75496 (None-None) Geminin
UniProt ID: Q9H211 (None-None) DNA replication factor Cdt1
|
|
|
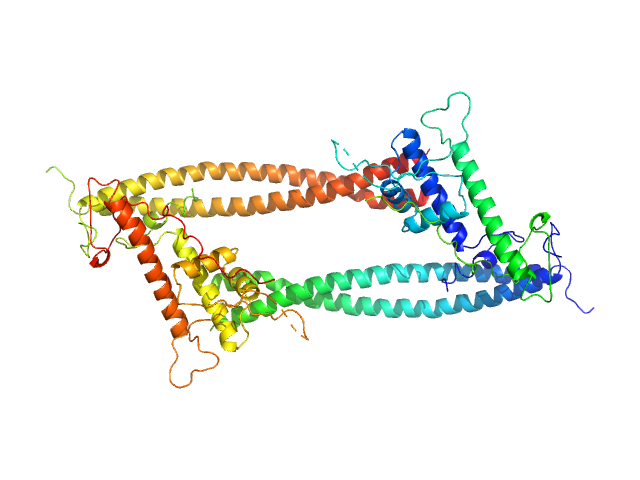
|
| Sample: |
Geminin dimer, 47 kDa Homo sapiens protein
DNA replication factor Cdt1 monomer, 60 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris75 200 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2006 Oct 5
|
Quaternary structure of the human Cdt1-Geminin complex regulates DNA replication licensing.
Proc Natl Acad Sci U S A 106(47):19807-12 (2009)
De Marco V, Gillespie PJ, Li A, Karantzelis N, Christodoulou E, Klompmaker R, van Gerwen S, Fish A, Petoukhov MV, Iliou MS, Lygerou Z, Medema RH, Blow JJ, Svergun DI, Taraviras S, Perrakis A
|
| RgGuinier |
3.8 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
120 |
nm3 |
|
|
UniProt ID: P23909 (None-None) DNA mismatch repair protein MutS
|
|
|
|
| Sample: |
DNA mismatch repair protein MutS tetramer, 381 kDa Escherichia coli protein
|
| Buffer: |
50 mM HEPES 50 mM KCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 May 12
|
Using stable MutS dimers and tetramers to quantitatively analyze DNA mismatch recognition and sliding clamp formation.
Nucleic Acids Res 41(17):8166-81 (2013)
Groothuizen FS, Fish A, Petoukhov MV, Reumer A, Manelyte L, Winterwerp HH, Marinus MG, Lebbink JH, Svergun DI, Friedhoff P, Sixma TK
|
| RgGuinier |
8.5 |
nm |
| Dmax |
29.0 |
nm |
| VolumePorod |
750 |
nm3 |
|
|
UniProt ID: P23909 (None-None) DNA mismatch repair protein MutS
|
|
|
|
| Sample: |
DNA mismatch repair protein MutS tetramer, 381 kDa Escherichia coli protein
|
| Buffer: |
50 mM HEPES 50 mM KCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 May 12
|
Using stable MutS dimers and tetramers to quantitatively analyze DNA mismatch recognition and sliding clamp formation.
Nucleic Acids Res 41(17):8166-81 (2013)
Groothuizen FS, Fish A, Petoukhov MV, Reumer A, Manelyte L, Winterwerp HH, Marinus MG, Lebbink JH, Svergun DI, Friedhoff P, Sixma TK
|
| RgGuinier |
8.3 |
nm |
| Dmax |
29.0 |
nm |
| VolumePorod |
720 |
nm3 |
|
|
UniProt ID: P23909 (None-None) DNA mismatch repair protein MutS
|
|
|
|
| Sample: |
DNA mismatch repair protein MutS tetramer, 381 kDa Escherichia coli protein
|
| Buffer: |
50 mM HEPES 50 mM KCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 May 12
|
Using stable MutS dimers and tetramers to quantitatively analyze DNA mismatch recognition and sliding clamp formation.
Nucleic Acids Res 41(17):8166-81 (2013)
Groothuizen FS, Fish A, Petoukhov MV, Reumer A, Manelyte L, Winterwerp HH, Marinus MG, Lebbink JH, Svergun DI, Friedhoff P, Sixma TK
|
|
|
UniProt ID: P23909 (None-None) DNA mismatch repair protein MutS
|
|
|
|
| Sample: |
DNA mismatch repair protein MutS tetramer, 381 kDa Escherichia coli protein
|
| Buffer: |
50 mM HEPES 50 mM KCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 May 12
|
Using stable MutS dimers and tetramers to quantitatively analyze DNA mismatch recognition and sliding clamp formation.
Nucleic Acids Res 41(17):8166-81 (2013)
Groothuizen FS, Fish A, Petoukhov MV, Reumer A, Manelyte L, Winterwerp HH, Marinus MG, Lebbink JH, Svergun DI, Friedhoff P, Sixma TK
|
| RgGuinier |
7.8 |
nm |
| Dmax |
27.0 |
nm |
|
|
UniProt ID: Q9VXJ0 (None-None) Peroxisomal multifunctional enzyme type 2
|
|
|
|
| Sample: |
Peroxisomal multifunctional enzyme type 2 dimer, 128 kDa Drosophila melanogaster protein
|
| Buffer: |
20 mM Sodium Phosphate 200 mM NaCl 5% (v/v) Glycerol 1mM Na2EDTA 1 mM NaN3, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2010 Jun 12
|
Quaternary structure of human, Drosophila melanogaster and Caenorhabditis elegans MFE-2 in solution from synchrotron small-angle X-ray scattering.
FEBS Lett 587(4):305-10 (2013)
Mehtälä ML, Haataja TJ, Blanchet CE, Hiltunen JK, Svergun DI, Glumoff T
|
| RgGuinier |
3.6 |
nm |
| Dmax |
12.0 |
nm |
|
|
UniProt ID: P51659 (None-None) Peroxisomal multifunctional enzyme type 2
|
|
|
|
| Sample: |
Peroxisomal multifunctional enzyme type 2 dimer, 159 kDa Homo sapiens protein
|
| Buffer: |
20 mM Sodium Phosphate 200 mM NaCl 5% (v/v) Glycerol 1mM Na2EDTA 1 mM NaN3, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Mar 22
|
Quaternary structure of human, Drosophila melanogaster and Caenorhabditis elegans MFE-2 in solution from synchrotron small-angle X-ray scattering.
FEBS Lett 587(4):305-10 (2013)
Mehtälä ML, Haataja TJ, Blanchet CE, Hiltunen JK, Svergun DI, Glumoff T
|
| RgGuinier |
4.6 |
nm |
| Dmax |
15.0 |
nm |
|
|
UniProt ID: Q6P5F9 (None-None) Exportin-1
|
|
|

|
| Sample: |
Exportin-1 monomer, 123 kDa Mus musculus protein
|
| Buffer: |
50 mM Tris-HCL 150 mM NaCl 1.0 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Feb 3
|
Structural determinants and mechanism of mammalian CRM1 allostery.
Structure 21(8):1350-60 (2013)
Dölker N, Blanchet CE, Voß B, Haselbach D, Kappel C, Monecke T, Svergun DI, Stark H, Ficner R, Zachariae U, Grubmüller H, Dickmanns A
|
| RgGuinier |
3.8 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
190 |
nm3 |
|
|
UniProt ID: Q6P5F9 (None-None) Exportin-1
UniProt ID: P62826 (None-None) GTP-binding nuclear protein Ran
|
|
|

|
| Sample: |
Exportin-1 monomer, 123 kDa Mus musculus protein
GTP-binding nuclear protein Ran monomer, 24 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris-HCL 150 mM NaCl 1.0 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Feb 3
|
Structural determinants and mechanism of mammalian CRM1 allostery.
Structure 21(8):1350-60 (2013)
Dölker N, Blanchet CE, Voß B, Haselbach D, Kappel C, Monecke T, Svergun DI, Stark H, Ficner R, Zachariae U, Grubmüller H, Dickmanns A
|
| RgGuinier |
3.6 |
nm |
| Dmax |
10.0 |
nm |
|
|
UniProt ID: Q6P5F9 (None-None) Exportin-1
UniProt ID: P62826 (None-None) GTP-binding nuclear protein Ran
UniProt ID: O95149 (None-None) Snurportin-1
|
|
|

|
| Sample: |
Exportin-1 monomer, 123 kDa Mus musculus protein
GTP-binding nuclear protein Ran monomer, 24 kDa Homo sapiens protein
Snurportin-1 monomer, 41 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris-HCL 150 mM NaCl 1.0 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Feb 3
|
Structural determinants and mechanism of mammalian CRM1 allostery.
Structure 21(8):1350-60 (2013)
Dölker N, Blanchet CE, Voß B, Haselbach D, Kappel C, Monecke T, Svergun DI, Stark H, Ficner R, Zachariae U, Grubmüller H, Dickmanns A
|
| RgGuinier |
4.1 |
nm |
| Dmax |
14.0 |
nm |
|
|