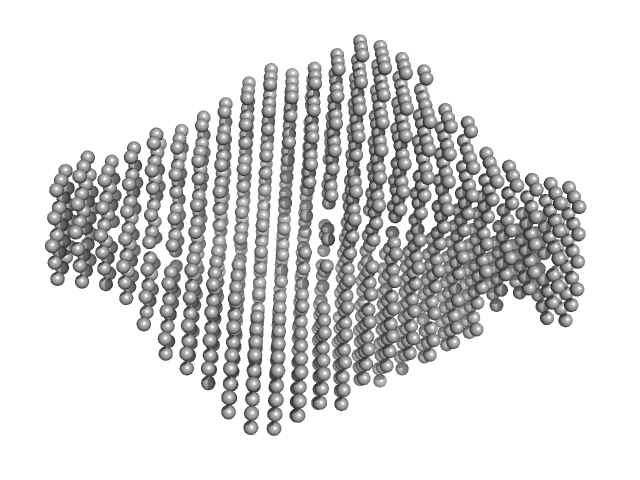
UniProt ID: Q6P5F9 (None-None) Exportin-1
UniProt ID: O95149 (None-None) Snurportin-1
|
|
|
|
| Sample: |
Exportin-1 monomer, 123 kDa Mus musculus protein
Snurportin-1 monomer, 41 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris-HCL 150 mM NaCl 1.0 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Dec 10
|
Structural determinants and mechanism of mammalian CRM1 allostery.
Structure 21(8):1350-60 (2013)
Dölker N, Blanchet CE, Voß B, Haselbach D, Kappel C, Monecke T, Svergun DI, Stark H, Ficner R, Zachariae U, Grubmüller H, Dickmanns A
|
| RgGuinier |
4.3 |
nm |
| Dmax |
15.0 |
nm |
|
|
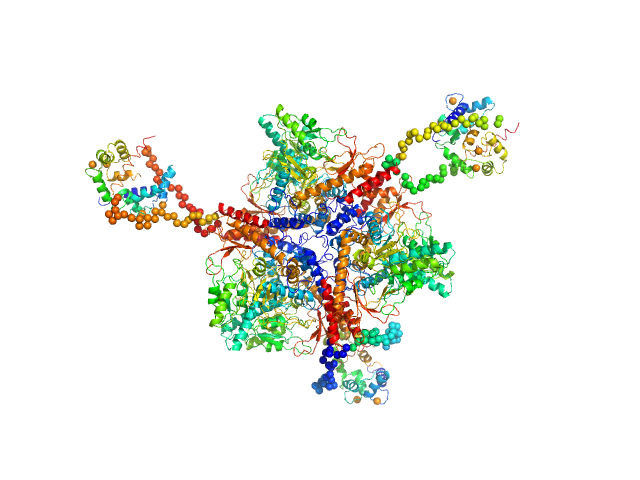
UniProt ID: P62158 (None-None) Calmodulin
UniProt ID: (None-None) C-terminal region of human myelin basic protein
|
|
|

|
| Sample: |
Calmodulin monomer, 17 kDa Homo sapiens protein
C-terminal region of human myelin basic protein monomer, 2 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris75 200 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2006 Nov 28
|
Interaction between the C-terminal region of human myelin basic protein and calmodulin: analysis of complex formation and solution structure.
BMC Struct Biol 8:10 (2008)
Majava V, Petoukhov MV, Hayashi N, Pirilä P, Svergun DI, Kursula P
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.0 |
nm |
| VolumePorod |
36 |
nm3 |
|
|
UniProt ID: B1VBB0 (None-None) Chitinase 60
|
|
|
|
| Sample: |
Chitinase 60 monomer, 61 kDa Moritella marina protein
|
| Buffer: |
20 mM Tris 200 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Mar 18
|
Crystal structures of substrate-bound chitinase from the psychrophilic bacterium Moritella marina and its structure in solution.
Acta Crystallogr D Biol Crystallogr 70(Pt 3):676-84 (2014)
Malecki PH, Vorgias CE, Petoukhov MV, Svergun DI, Rypniewski W
|
| RgGuinier |
3.2 |
nm |
| Dmax |
11.3 |
nm |
| VolumePorod |
75 |
nm3 |
|
|
UniProt ID: P62158 (None-None) Calmodulin
UniProt ID: Q07346 (None-None) Glutamate decarboxylase
|
|
|
|
| Sample: |
Calmodulin monomer, 17 kDa Homo sapiens protein
Glutamate decarboxylase hexamer, 340 kDa Petunia x hybrida protein
|
| Buffer: |
50 mM HEPES 50 mM KCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2004 Nov 9
|
A common structural basis for pH- and calmodulin-mediated regulation in plant glutamate decarboxylase.
J Mol Biol 392(2):334-51 (2009)
Gut H, Dominici P, Pilati S, Astegno A, Petoukhov MV, Svergun DI, Grütter MG, Capitani G
|
| RgGuinier |
5.9 |
nm |
| Dmax |
23.0 |
nm |
| VolumePorod |
630 |
nm3 |
|
|
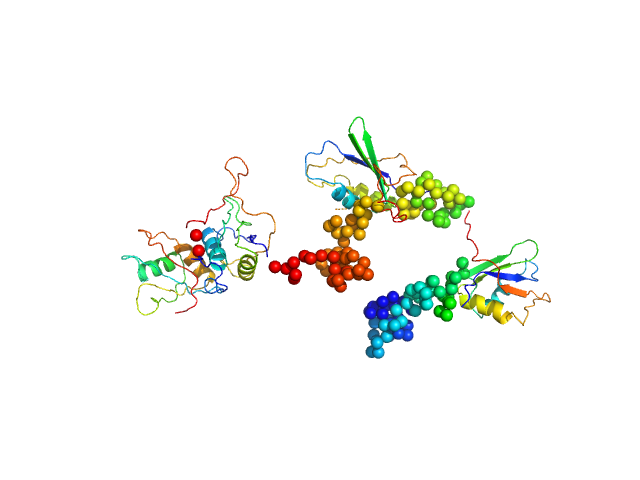
UniProt ID: P26599 (None-None) Polypyrimidine tract-binding protein 1
|
|
|
|
| Sample: |
Polypyrimidine tract-binding protein 1 monomer, 57 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris 100 mM NaCl 5.0 mM DTT, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2004 Feb 11
|
Conformation of polypyrimidine tract binding protein in solution.
Structure 14(6):1021-7 (2006)
Petoukhov MV, Monie TP, Allain FH, Matthews S, Curry S, Svergun DI
|
| RgGuinier |
4.0 |
nm |
| Dmax |
16.5 |
nm |
|
|
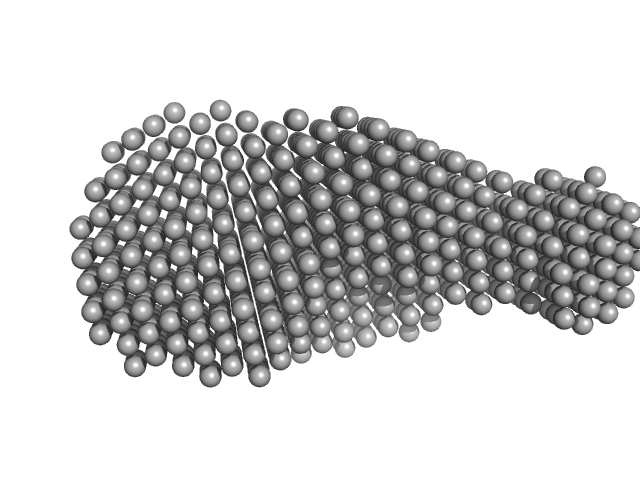
UniProt ID: Q03405 (None-None) Urokinase plasminogen activator surface receptor
|
|
|
|
| Sample: |
Urokinase plasminogen activator surface receptor monomer, 37 kDa Homo sapiens protein
|
| Buffer: |
25 mM Sodium Phosphate 5 % Glycerol 50 mM NaSO4, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2010 Jun 10
|
A flexible multidomain structure drives the function of the urokinase-type plasminogen activator receptor (uPAR).
J Biol Chem 287(41):34304-15 (2012)
Mertens HD, Kjaergaard M, Mysling S, Gårdsvoll H, Jørgensen TJ, Svergun DI, Ploug M
|
| RgGuinier |
2.8 |
nm |
| Dmax |
9.7 |
nm |
| VolumePorod |
69 |
nm3 |
|
|
UniProt ID: Q03405 (None-None) Urokinase plasminogen activator surface receptor
|
|
|

|
| Sample: |
Urokinase plasminogen activator surface receptor monomer, 37 kDa Homo sapiens protein
|
| Buffer: |
25 mM Sodium Phosphate 5 % Glycerol 50 mM NaSO4, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2010 Jun 10
|
A flexible multidomain structure drives the function of the urokinase-type plasminogen activator receptor (uPAR).
J Biol Chem 287(41):34304-15 (2012)
Mertens HD, Kjaergaard M, Mysling S, Gårdsvoll H, Jørgensen TJ, Svergun DI, Ploug M
|
| RgGuinier |
2.2 |
nm |
| Dmax |
7.0 |
nm |
| VolumePorod |
57 |
nm3 |
|
|
UniProt ID: Q03405 (None-None) Urokinase plasminogen activator surface receptor
UniProt ID: (None-None) synthetic peptide AE105
|
|
|
|
| Sample: |
Urokinase plasminogen activator surface receptor monomer, 37 kDa Homo sapiens protein
Synthetic peptide AE105 monomer, 1 kDa protein
|
| Buffer: |
25 mM Sodium Phosphate 5 % Glycerol 50 mM NaSO4, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2010 Jun 10
|
A flexible multidomain structure drives the function of the urokinase-type plasminogen activator receptor (uPAR).
J Biol Chem 287(41):34304-15 (2012)
Mertens HD, Kjaergaard M, Mysling S, Gårdsvoll H, Jørgensen TJ, Svergun DI, Ploug M
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
61 |
nm3 |
|
|
UniProt ID: Q03405 (None-None) Urokinase plasminogen activator surface receptor
UniProt ID: (None-None) synthetic peptide AE234
|
|
|
|
| Sample: |
Urokinase plasminogen activator surface receptor monomer, 37 kDa Homo sapiens protein
Synthetic peptide AE234 monomer, 1 kDa protein
|
| Buffer: |
25 mM Sodium Phosphate 5 % Glycerol 50 mM NaSO4, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2010 Jun 10
|
A flexible multidomain structure drives the function of the urokinase-type plasminogen activator receptor (uPAR).
J Biol Chem 287(41):34304-15 (2012)
Mertens HD, Kjaergaard M, Mysling S, Gårdsvoll H, Jørgensen TJ, Svergun DI, Ploug M
|
| RgGuinier |
2.4 |
nm |
| Dmax |
8.3 |
nm |
| VolumePorod |
57 |
nm3 |
|
|
UniProt ID: Q03405 (None-None) Urokinase plasminogen activator surface receptor
UniProt ID: P00749 (None-None) Urokinase-type plasminogen activator
|
|
|
|
| Sample: |
Urokinase plasminogen activator surface receptor monomer, 37 kDa Homo sapiens protein
Urokinase-type plasminogen activator monomer, 49 kDa Homo sapiens protein
|
| Buffer: |
25 mM Sodium Phosphate 5 % Glycerol 50 mM NaSO4, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2010 Jun 10
|
A flexible multidomain structure drives the function of the urokinase-type plasminogen activator receptor (uPAR).
J Biol Chem 287(41):34304-15 (2012)
Mertens HD, Kjaergaard M, Mysling S, Gårdsvoll H, Jørgensen TJ, Svergun DI, Ploug M
|
| RgGuinier |
2.8 |
nm |
| Dmax |
9.4 |
nm |
| VolumePorod |
102 |
nm3 |
|
|