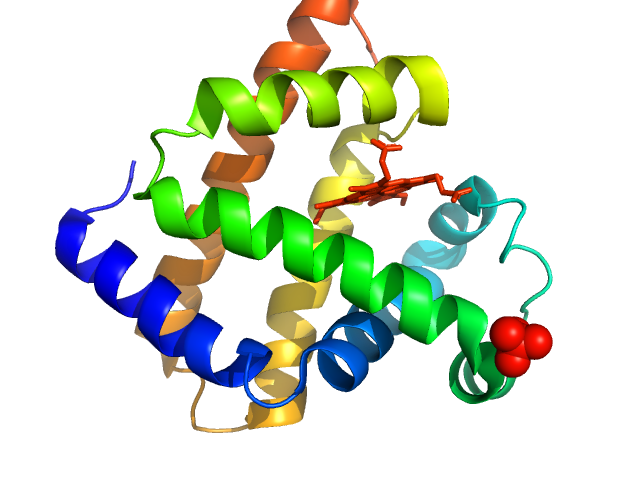
UniProt ID: P68082 (None-None) Myoglobin
|
|
|
|
| Sample: |
Myoglobin monomer, 17 kDa Equus caballus protein
|
| Buffer: |
PBS, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2012 Sep 20
|
Standard proteins
Darja Ruskule
|
| RgGuinier |
1.6 |
nm |
| Dmax |
5.0 |
nm |
| VolumePorod |
32 |
nm3 |
|
|
UniProt ID: P01012 (None-None) Ovalbumin
|
|
|

|
| Sample: |
Ovalbumin monomer, 43 kDa Gallus gallus protein
|
| Buffer: |
PBS, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2012 Sep 18
|
Standard proteins
Darja Ruskule
|
| RgGuinier |
2.5 |
nm |
| Dmax |
7.8 |
nm |
| VolumePorod |
74 |
nm3 |
|
|
UniProt ID: P61823 (None-None) Ribonuclease pancreatic
|
|
|
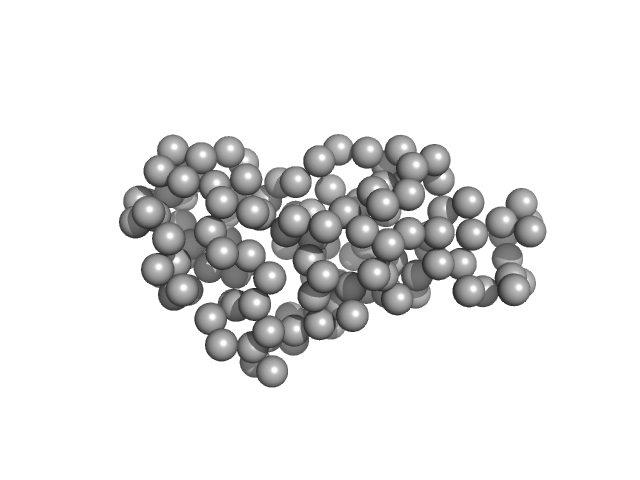
|
| Sample: |
Ribonuclease pancreatic monomer, 16 kDa Bos taurus protein
|
| Buffer: |
PBS, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2012 Sep 18
|
Standard proteins
Darja Ruskule
|
| RgGuinier |
1.6 |
nm |
| Dmax |
5.0 |
nm |
| VolumePorod |
15 |
nm3 |
|
|
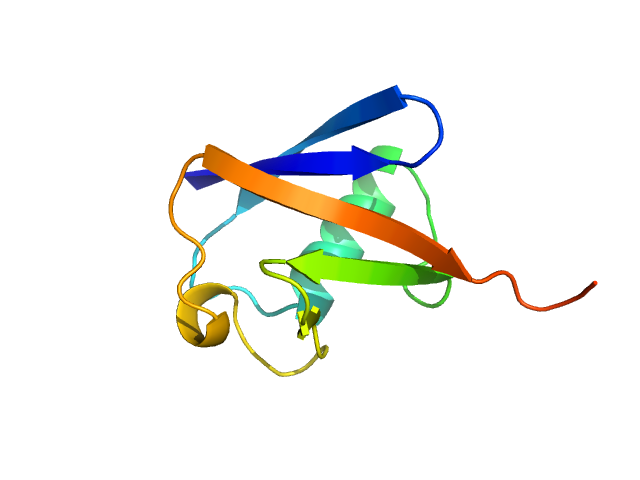
UniProt ID: P63048 (1-76) Ubiquitin
|
|
|
|
| Sample: |
Ubiquitin monomer, 9 kDa Bos taurus protein
|
| Buffer: |
40 mM Sodium acetate 150 mM NaCl, pH: 5.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2012 Sep 18
|
Standard proteins
Darja Ruskule
|
| RgGuinier |
1.3 |
nm |
| Dmax |
4.9 |
nm |
| VolumePorod |
12 |
nm3 |
|
|
UniProt ID: P61823 (None-None) Ribonuclease pancreatic
|
|
|

|
| Sample: |
Ribonuclease pancreatic monomer, 16 kDa Bos taurus protein
|
| Buffer: |
PBS, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2012 Sep 18
|
Standard proteins
Darja Ruskule
|
| RgGuinier |
1.6 |
nm |
| Dmax |
5.0 |
nm |
| VolumePorod |
17 |
nm3 |
|
|
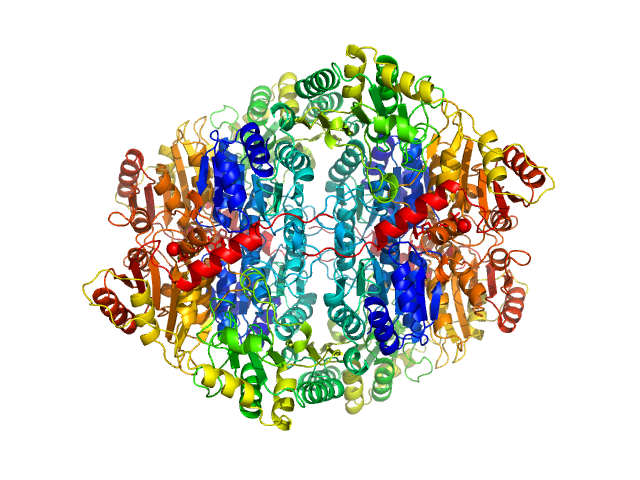
UniProt ID: P06672 (None-None) Pyruvate decarboxylase
|
|
|
|
| Sample: |
Pyruvate decarboxylase tetramer, 244 kDa Zymomonas mobilis protein
|
| Buffer: |
100 mM Sodium Citrate, 17% Glycerol, 22.5% PEG 1500, pH: 6 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 1998 Nov 3
|
Crystal versus solution structures of thiamine diphosphate-dependent enzymes.
J Biol Chem 275(1):297-302 (2000)
Svergun DI, Petoukhov MV, Koch MH, König S
|
| RgGuinier |
3.9 |
nm |
| Dmax |
11.0 |
nm |
|
|
UniProt ID: P23909 (None-None) DNA mismatch repair protein MutS
|
|
|

|
| Sample: |
DNA mismatch repair protein MutS dimer, 191 kDa Escherichia coli protein
|
| Buffer: |
50 mM HEPES 50 mM KCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Feb 28
|
Using stable MutS dimers and tetramers to quantitatively analyze DNA mismatch recognition and sliding clamp formation.
Nucleic Acids Res 41(17):8166-81 (2013)
Groothuizen FS, Fish A, Petoukhov MV, Reumer A, Manelyte L, Winterwerp HH, Marinus MG, Lebbink JH, Svergun DI, Friedhoff P, Sixma TK
|
| RgGuinier |
4.7 |
nm |
| Dmax |
15.5 |
nm |
| VolumePorod |
307 |
nm3 |
|
|
UniProt ID: P23909 (None-None) DNA mismatch repair protein MutS
|
|
|
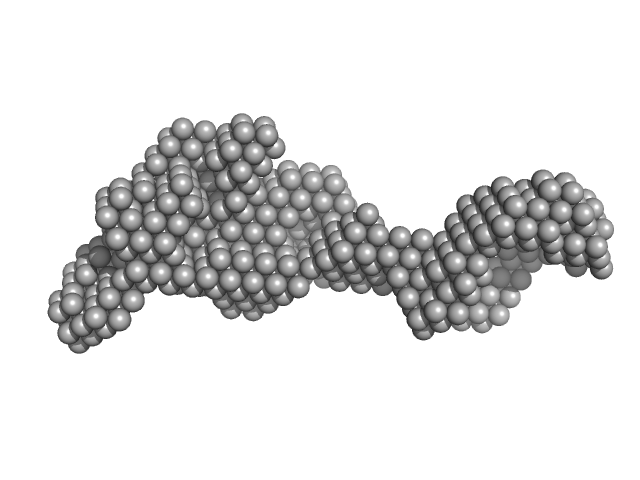
|
| Sample: |
DNA mismatch repair protein MutS tetramer, 381 kDa Escherichia coli protein
|
| Buffer: |
50 mM HEPES 50 mM KCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 May 12
|
Using stable MutS dimers and tetramers to quantitatively analyze DNA mismatch recognition and sliding clamp formation.
Nucleic Acids Res 41(17):8166-81 (2013)
Groothuizen FS, Fish A, Petoukhov MV, Reumer A, Manelyte L, Winterwerp HH, Marinus MG, Lebbink JH, Svergun DI, Friedhoff P, Sixma TK
|
| RgGuinier |
7.8 |
nm |
| Dmax |
28.0 |
nm |
| VolumePorod |
700 |
nm3 |
|
|
UniProt ID: A0A0H2URK1 (187-385) Functional binding region (187-385) of the pneumococcal serine-rich repeat protein
|
|
|
|
| Sample: |
Functional binding region (187-385) of the pneumococcal serine-rich repeat protein monomer, 22 kDa Streptococcus pneumoniae protein
|
| Buffer: |
20 mM sodium citrate 250 mM NaCl 2.5 % Glycerol, pH: 5.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Jul 2
|
The basic keratin 10-binding domain of the virulence-associated pneumococcal serine-rich protein PsrP adopts a novel MSCRAMM fold.
Open Biol 4:130090 (2014)
Schulte T, Löfling J, Mikaelsson C, Kikhney A, Hentrich K, Diamante A, Ebel C, Normark S, Svergun D, Henriques-Normark B, Achour A
|
| RgGuinier |
2.3 |
nm |
| Dmax |
7.7 |
nm |
| VolumePorod |
37 |
nm3 |
|
|
UniProt ID: O75496 (None-None) Geminin
UniProt ID: Q9H211 (None-None) DNA replication factor Cdt1
|
|
|
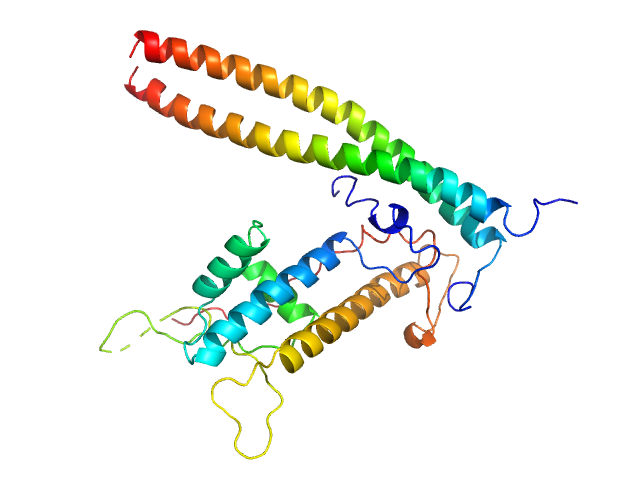
|
| Sample: |
Geminin dimer, 47 kDa Homo sapiens protein
DNA replication factor Cdt1 monomer, 60 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris75 200 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2006 Aug 13
|
Quaternary structure of the human Cdt1-Geminin complex regulates DNA replication licensing.
Proc Natl Acad Sci U S A 106(47):19807-12 (2009)
De Marco V, Gillespie PJ, Li A, Karantzelis N, Christodoulou E, Klompmaker R, van Gerwen S, Fish A, Petoukhov MV, Iliou MS, Lygerou Z, Medema RH, Blow JJ, Svergun DI, Taraviras S, Perrakis A
|
| RgGuinier |
2.9 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
70 |
nm3 |
|
|