UniProt ID: Q6CSX2 (562-831) Serine/threonine-protein kinase ATG1
UniProt ID: Q6CWK2 (400-475) Autophagy-related protein 13
UniProt ID: Q6CS99 (1-423) Autophagy-related protein 17
UniProt ID: Q6CTU8 (1-85) Autophagy-related protein 29
UniProt ID: Q6CX74 (1-143) KLLA0A10637p
|
|
|
|
| Sample: |
Serine/threonine-protein kinase ATG1 dimer, 61 kDa Kluyveromyces lactis protein
Autophagy-related protein 13 dimer, 17 kDa Kluyveromyces lactis protein
Autophagy-related protein 17 dimer, 100 kDa Kluyveromyces lactis protein
Autophagy-related protein 29 dimer, 20 kDa Kluyveromyces lactis protein
KLLA0A10637p dimer, 32 kDa Kluyveromyces lactis protein
|
| Buffer: |
20 mM Tris, 200 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2013 Dec 20
|
Solution structure of the Atg1 complex: implications for the architecture of the phagophore assembly site.
Structure 23(5):809-818 (2015)
Köfinger J, Ragusa MJ, Lee IH, Hummer G, Hurley JH
|
| RgGuinier |
10.3 |
nm |
| Dmax |
34.0 |
nm |
| VolumePorod |
1000 |
nm3 |
|
|
UniProt ID: Q07812 (1-192) Apoptosis regulator BAX (Bcl-2 associated X)
|
|
|
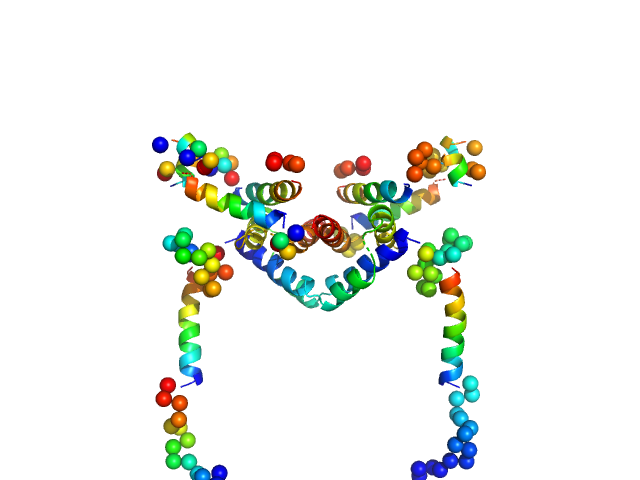
|
| Sample: |
Apoptosis regulator BAX (Bcl-2 associated X) dimer, 42 kDa Homo sapiens protein
|
| Buffer: |
20mM sodium phosphate 100mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at 23A, Taiwan Photon Source, NSRRC on 2015 May 28
|
Oligomerization process of Bcl-2 associated X protein revealed from intermediate structures in solution.
Phys Chem Chem Phys 19(11):7947-7954 (2017)
Shih O, Yeh YQ, Liao KF, Sung TC, Chiang YW, Jeng US
|
| RgGuinier |
3.0 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
86 |
nm3 |
|
|
UniProt ID: Q07812 (1-192) Apoptosis regulator BAX (Bcl-2 associated X)
|
|
|
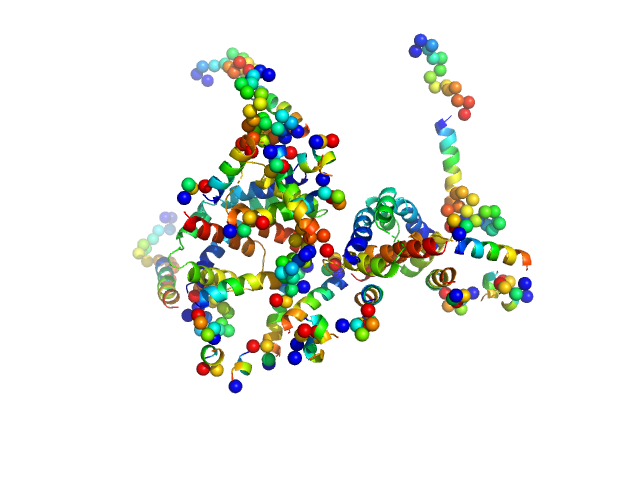
|
| Sample: |
Apoptosis regulator BAX (Bcl-2 associated X) tetramer, 85 kDa Homo sapiens protein
|
| Buffer: |
20mM sodium phosphate 100mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at 23A, Taiwan Photon Source, NSRRC on 2015 May 28
|
Oligomerization process of Bcl-2 associated X protein revealed from intermediate structures in solution.
Phys Chem Chem Phys 19(11):7947-7954 (2017)
Shih O, Yeh YQ, Liao KF, Sung TC, Chiang YW, Jeng US
|
| RgGuinier |
3.6 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
180 |
nm3 |
|
|
UniProt ID: Q8NAP3 (1006-1152) Zinc finger and BTB domain-containing protein 38
UniProt ID: (None-None) methylated C-terminal ZBTB38 binding sequence
|
|
|

|
| Sample: |
Zinc finger and BTB domain-containing protein 38 monomer, 17 kDa Homo sapiens protein
Methylated C-terminal ZBTB38 binding sequence monomer, 17 kDa DNA
|
| Buffer: |
10 mM Tris, 1 mM tris(2-carboxy-ethyl)phosphine (TCEP), 0.05% NaN3, 10% D2O, pH: 6.8 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSess, Department of Chemistry, University of Utah on 2016 Nov 11
|
The C-Terminal Zinc Fingers of ZBTB38 are Novel Selective Readers of DNA Methylation.
J Mol Biol 430(3):258-271 (2018)
Pozner A, Hudson NO, Trewhella J, Terooatea TW, Miller SA, Buck-Koehntop BA
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.2 |
nm |
| VolumePorod |
50 |
nm3 |
|
|
UniProt ID: P31483-2 (None-None) Nucleolysin TIA-1 isoform p40
UniProt ID: (None-None) poly U 15mer
|
|
|
|
| Sample: |
Nucleolysin TIA-1 isoform p40 monomer, 30 kDa Homo sapiens protein
Poly U 15mer monomer, 5 kDa RNA
|
| Buffer: |
10 mM Potassium Phosphate 50 mM NaCl 10 mM DTT, pH: 6 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, SFB 1035, Technische Universität München on 2016 Apr 28
|
Segmental, Domain-Selective Perdeuteration and Small-Angle Neutron Scattering for Structural Analysis of Multi-Domain Proteins.
Angew Chem Int Ed Engl 56(32):9322-9325 (2017)
Sonntag M, Jagtap PKA, Simon B, Appavou MS, Geerlof A, Stehle R, Gabel F, Hennig J, Sattler M
|
| RgGuinier |
2.4 |
nm |
| Dmax |
8.7 |
nm |
| VolumePorod |
39 |
nm3 |
|
|
UniProt ID: P31483-2 (None-None) Nucleolysin TIA-1 isoform p40
|
|
|
|
| Sample: |
Nucleolysin TIA-1 isoform p40 monomer, 31 kDa Homo sapiens protein
|
| Buffer: |
10 mM Potassium Phosphate 50 mM NaCl 10 mM DTT, pH: 6 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, SFB 1035, Technische Universität München on 2015 Dec 1
|
Segmental, Domain-Selective Perdeuteration and Small-Angle Neutron Scattering for Structural Analysis of Multi-Domain Proteins.
Angew Chem Int Ed Engl 56(32):9322-9325 (2017)
Sonntag M, Jagtap PKA, Simon B, Appavou MS, Geerlof A, Stehle R, Gabel F, Hennig J, Sattler M
|
| RgGuinier |
2.7 |
nm |
| Dmax |
10.3 |
nm |
| VolumePorod |
38 |
nm3 |
|
|
UniProt ID: (None-None) poly U 15mer
UniProt ID: P31483-2 (None-None) Nucleolysin TIA-1 isoform p40
|
|
|
|
| Sample: |
Poly U 15mer monomer, 5 kDa RNA
Nucleolysin TIA-1 isoform p40 monomer, 31 kDa Homo sapiens protein
|
| Buffer: |
10 mM Potassium Phosphate 50 mM NaCl 10 mM DTT, pH: 6 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, SFB 1035, Technische Universität München on 2015 Dec 1
|
Segmental, Domain-Selective Perdeuteration and Small-Angle Neutron Scattering for Structural Analysis of Multi-Domain Proteins.
Angew Chem Int Ed Engl 56(32):9322-9325 (2017)
Sonntag M, Jagtap PKA, Simon B, Appavou MS, Geerlof A, Stehle R, Gabel F, Hennig J, Sattler M
|
| RgGuinier |
2.4 |
nm |
| Dmax |
8.6 |
nm |
| VolumePorod |
40 |
nm3 |
|
|
UniProt ID: P31483-2 (None-None) Nucleolysin TIA-1 isoform p40
|
|
|
|
| Sample: |
Nucleolysin TIA-1 isoform p40 monomer, 30 kDa Homo sapiens protein
|
| Buffer: |
10 mM Potassium Phosphate 50 mM NaCl 10 mM DTT, pH: 6 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, SFB 1035, Technische Universität München on 2016 Apr 28
|
Segmental, Domain-Selective Perdeuteration and Small-Angle Neutron Scattering for Structural Analysis of Multi-Domain Proteins.
Angew Chem Int Ed Engl 56(32):9322-9325 (2017)
Sonntag M, Jagtap PKA, Simon B, Appavou MS, Geerlof A, Stehle R, Gabel F, Hennig J, Sattler M
|
| RgGuinier |
2.7 |
nm |
| Dmax |
11.2 |
nm |
| VolumePorod |
38 |
nm3 |
|
|
UniProt ID: P31483-2 (None-None) Nucleolysin TIA-1 isoform p40
|
|
|
|
| Sample: |
Nucleolysin TIA-1 isoform p40 monomer, 30 kDa Homo sapiens protein
|
| Buffer: |
10 mM Potassium Phosphate 50 mM NaCl 10 mM DTT, pH: 6 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, SFB 1035, Technische Universität München on 2016 May 4
|
Segmental, Domain-Selective Perdeuteration and Small-Angle Neutron Scattering for Structural Analysis of Multi-Domain Proteins.
Angew Chem Int Ed Engl 56(32):9322-9325 (2017)
Sonntag M, Jagtap PKA, Simon B, Appavou MS, Geerlof A, Stehle R, Gabel F, Hennig J, Sattler M
|
| RgGuinier |
2.7 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
37 |
nm3 |
|
|
UniProt ID: (None-None) poly U 15mer
UniProt ID: P31483-2 (None-None) Nucleolysin TIA-1 isoform p40
|
|
|
|
| Sample: |
Poly U 15mer monomer, 5 kDa RNA
Nucleolysin TIA-1 isoform p40 monomer, 30 kDa Homo sapiens protein
|
| Buffer: |
10 mM Potassium Phosphate 50 mM NaCl 10 mM DTT, pH: 6 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, SFB 1035, Technische Universität München on 2016 May 4
|
Segmental, Domain-Selective Perdeuteration and Small-Angle Neutron Scattering for Structural Analysis of Multi-Domain Proteins.
Angew Chem Int Ed Engl 56(32):9322-9325 (2017)
Sonntag M, Jagtap PKA, Simon B, Appavou MS, Geerlof A, Stehle R, Gabel F, Hennig J, Sattler M
|
| RgGuinier |
2.4 |
nm |
| Dmax |
8.6 |
nm |
| VolumePorod |
40 |
nm3 |
|
|