UniProt ID: P02769 (25-607) Serum albumin
|
|
|

|
| Sample: |
Serum albumin monomer, 66 kDa Bos taurus protein
|
| Buffer: |
25 mM MOPS, 250 mM NaCl, 50 mM KCl, 2 mM TCEP, 0.1% NaN3, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2017 Mar 9
|
2017 publication guidelines for structural modelling of small-angle scattering data from biomolecules in solution: an update.
Acta Crystallogr D Struct Biol 73(Pt 9):710-728 (2017)
Trewhella J, Duff AP, Durand D, Gabel F, Guss JM, Hendrickson WA, Hura GL, Jacques DA, Kirby NM, Kwan AH, Pérez J, Pollack L, Ryan TM, Sali A, Schneidman-Duhovny D, Schwede T, Svergun DI, Sugiyama M, Tainer JA, Vachette P, Westbrook J, Whitten AE
|
| RgGuinier |
2.8 |
nm |
| Dmax |
8.7 |
nm |
| VolumePorod |
101 |
nm3 |
|
|
UniProt ID: Q6CSX2 (562-831) Serine/threonine-protein kinase ATG1
UniProt ID: Q6CWK2 (400-475) Autophagy-related protein 13
|
|
|
|
| Sample: |
Serine/threonine-protein kinase ATG1 dimer, 61 kDa Kluyveromyces lactis protein
Autophagy-related protein 13 dimer, 17 kDa Kluyveromyces lactis protein
|
| Buffer: |
20 mM Tris, 200 mM NaCl, 2% glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2013 Dec 10
|
Solution structure of the Atg1 complex: implications for the architecture of the phagophore assembly site.
Structure 23(5):809-818 (2015)
Köfinger J, Ragusa MJ, Lee IH, Hummer G, Hurley JH
|
| RgGuinier |
3.3 |
nm |
| Dmax |
10.9 |
nm |
| VolumePorod |
160 |
nm3 |
|
|
UniProt ID: Q6CS99 (1-423) Autophagy-related protein 17
UniProt ID: Q6CTU8 (1-85) Autophagy-related protein 29
UniProt ID: Q6CX74 (1-143) KLLA0A10637p
|
|
|
|
| Sample: |
Autophagy-related protein 17 dimer, 100 kDa Kluyveromyces lactis protein
Autophagy-related protein 29 dimer, 20 kDa Kluyveromyces lactis protein
KLLA0A10637p dimer, 32 kDa Kluyveromyces lactis protein
|
| Buffer: |
20 mM Tris, 200 mM NaCl, 2% glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2013 Dec 10
|
Solution structure of the Atg1 complex: implications for the architecture of the phagophore assembly site.
Structure 23(5):809-818 (2015)
Köfinger J, Ragusa MJ, Lee IH, Hummer G, Hurley JH
|
| RgGuinier |
10.1 |
nm |
| Dmax |
34.0 |
nm |
| VolumePorod |
848 |
nm3 |
|
|
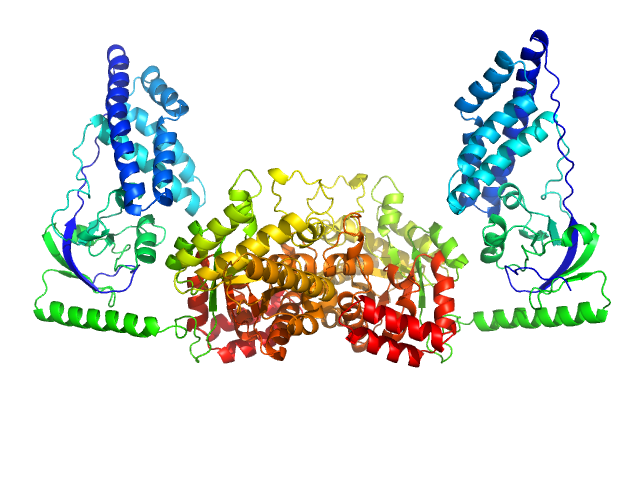
UniProt ID: A0A037YGN3 (1-575) Phosphoenolpyruvate-protein phosphotransferase
|
|
|
|
| Sample: |
Phosphoenolpyruvate-protein phosphotransferase dimer, 127 kDa Escherichia coli protein
|
| Buffer: |
20mM TRIS buffer, 100 mM NaCl, 10 mM DTT, 4 mM MgCl2, 1 mM EDTA, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12-ID-C, Advanced Photon Source (APS), Argonne National Laboratory on 2010 Aug 23
|
Solution structure of the 128 kDa enzyme I dimer from Escherichia coli and its 146 kDa complex with HPr using residual dipolar couplings and small- and wide-angle X-ray scattering.
J Am Chem Soc 132(37):13026-45 (2010)
Schwieters CD, Suh JY, Grishaev A, Ghirlando R, Takayama Y, Clore GM
|
| RgGuinier |
4.1 |
nm |
| Dmax |
14.8 |
nm |
| VolumePorod |
189 |
nm3 |
|
|
UniProt ID: Q6MNK1 (None-None) Bifunctional protein PutA
|
|
|

|
| Sample: |
Bifunctional protein PutA dimer, 219 kDa Bdellovibrio bacteriovorus protein
|
| Buffer: |
50 mM Tris, 125 mM NaCl, 1 mM EDTA, and 1 mM tris(3-hydroxypropyl)phosphine (THP) at pH 7.5,, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2012 Jun 8
|
Biophysical investigation of type A PutAs reveals a conserved core oligomeric structure.
FEBS J 284(18):3029-3049 (2017)
Korasick DA, Singh H, Pemberton TA, Luo M, Dhatwalia R, Tanner JJ
|
| RgGuinier |
4.5 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
287 |
nm3 |
|
|
UniProt ID: A0A0E0T6R2 (None-None) Bifunctional protein PutA
|
|
|
|
| Sample: |
Bifunctional protein PutA dimer, 229 kDa Desulfovibrio vulgaris protein
|
| Buffer: |
50 mM Tris-HCl, 50 mM NaCl, 0.5 mM EDTA, and 0.5 mM THP at pH 7.5., pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2012 Jun 8
|
Biophysical investigation of type A PutAs reveals a conserved core oligomeric structure.
FEBS J 284(18):3029-3049 (2017)
Korasick DA, Singh H, Pemberton TA, Luo M, Dhatwalia R, Tanner JJ
|
| RgGuinier |
4.4 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
293 |
nm3 |
|
|
UniProt ID: Q5ZUU6 (None-None) Bifunctional protein PutA
|
|
|
|
| Sample: |
Bifunctional protein PutA dimer, 238 kDa Legionella pneumophila subsp. … protein
|
| Buffer: |
50 mM Tris-HCl, 50 mM NaCl, 0.5 mM EDTA, and 0.5 mM THP at pH 7.5., pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2010 Apr 20
|
Biophysical investigation of type A PutAs reveals a conserved core oligomeric structure.
FEBS J 284(18):3029-3049 (2017)
Korasick DA, Singh H, Pemberton TA, Luo M, Dhatwalia R, Tanner JJ
|
| RgGuinier |
4.6 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
291 |
nm3 |
|
|
UniProt ID: Q89E26 (None-None) Proline dehydrogenase
|
|
|
|
| Sample: |
Proline dehydrogenase tetramer, 430 kDa Bradyrhizobium diazoefficiens protein
|
| Buffer: |
50 mM Tris (pH 7.8), 50 mM NaCl, 0.5 mM Tris(2-carboxyethyl)phosphine, and 5% (v/v) glycerol, pH: 7.8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2016 Dec 16
|
Biophysical investigation of type A PutAs reveals a conserved core oligomeric structure.
FEBS J 284(18):3029-3049 (2017)
Korasick DA, Singh H, Pemberton TA, Luo M, Dhatwalia R, Tanner JJ
|
| RgGuinier |
5.3 |
nm |
| Dmax |
14.1 |
nm |
| VolumePorod |
541 |
nm3 |
|
|
UniProt ID: Q89E26 (None-None) Proline dehydrogenase
|
|
|
|
| Sample: |
Proline dehydrogenase tetramer, 430 kDa Bradyrhizobium diazoefficiens protein
|
| Buffer: |
50 mM Tris (pH 7.8), 50 mM NaCl, 0.5 mM Tris(2-carboxyethyl)phosphine, and 5% (v/v) glycerol, pH: 7.8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2016 Dec 12
|
Biophysical investigation of type A PutAs reveals a conserved core oligomeric structure.
FEBS J 284(18):3029-3049 (2017)
Korasick DA, Singh H, Pemberton TA, Luo M, Dhatwalia R, Tanner JJ
|
| RgGuinier |
5.2 |
nm |
| Dmax |
14.6 |
nm |
| VolumePorod |
553 |
nm3 |
|
|
UniProt ID: Q89E26 (None-None) Proline dehydrogenase
|
|
|

|
| Sample: |
Proline dehydrogenase tetramer, 430 kDa Bradyrhizobium diazoefficiens protein
|
| Buffer: |
50 mM Tris (pH 7.8), 50 mM NaCl, 0.5 mM Tris(2-carboxyethyl)phosphine, and 5% (v/v) glycerol, pH: 7.8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2016 Dec 12
|
Biophysical investigation of type A PutAs reveals a conserved core oligomeric structure.
FEBS J 284(18):3029-3049 (2017)
Korasick DA, Singh H, Pemberton TA, Luo M, Dhatwalia R, Tanner JJ
|
| RgGuinier |
5.2 |
nm |
| Dmax |
13.7 |
nm |
| VolumePorod |
560 |
nm3 |
|
|