UniProt ID: P10636-8 (244-372) Isoform Tau-F of Microtubule-associated protein tau (C291A, K311C, K317C, C322A)
UniProt ID: A0A091E260 (1-77) Polyubiquitin-B
|
|
|
|
| Sample: |
Isoform Tau-F of Microtubule-associated protein tau (C291A, K311C, K317C, C322A) monomer, 14 kDa Homo sapiens protein
Polyubiquitin-B monomer, 9 kDa Fukomys damarensis protein
|
| Buffer: |
100 mM NaCl, pH: 6.8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2023 Mar 17
|
Conformational signatures induced by ubiquitin modification in the amyloid-forming tau repeat domain.
Proc Natl Acad Sci U S A 122(15):e2425831122 (2025)
Viola G, Trivellato D, Laitaoja M, Jänis J, Felli IC, D'Onofrio M, Mollica L, Giachin G, Assfalg M
|
| RgGuinier |
3.1 |
nm |
| Dmax |
12.1 |
nm |
| VolumePorod |
33 |
nm3 |
|
|
UniProt ID: P10636-8 (244-372) Isoform Tau-F of Microtubule-associated protein tau (C291A, K311C, K317C, C322A)
UniProt ID: A0A091E260 (1-77) Polyubiquitin-B
UniProt ID: A0A091E260 (1-77) Polyubiquitin-B
|
|
|
|
| Sample: |
Isoform Tau-F of Microtubule-associated protein tau (C291A, K311C, K317C, C322A) monomer, 14 kDa Homo sapiens protein
Polyubiquitin-B monomer, 9 kDa Fukomys damarensis protein
Polyubiquitin-B monomer, 9 kDa Fukomys damarensis protein
|
| Buffer: |
100 mM NaCl, pH: 6.8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2023 Mar 17
|
Conformational signatures induced by ubiquitin modification in the amyloid-forming tau repeat domain.
Proc Natl Acad Sci U S A 122(15):e2425831122 (2025)
Viola G, Trivellato D, Laitaoja M, Jänis J, Felli IC, D'Onofrio M, Mollica L, Giachin G, Assfalg M
|
| RgGuinier |
3.3 |
nm |
| Dmax |
12.2 |
nm |
| VolumePorod |
59 |
nm3 |
|
|
UniProt ID: P26022 (18-381) Pentraxin-related protein PTX3
|
|
|

|
| Sample: |
Pentraxin-related protein PTX3 octamer, 321 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris-HCl, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Sep 28
|
The structural organisation of pentraxin-3 and its interactions with heavy chains of inter-α-inhibitor regulate crosslinking of the hyaluronan matrix
Matrix Biology 136:52-68 (2025)
Shah A, Zhang X, Snee M, Lockhart-Cairns M, Levy C, Jowitt T, Birchenough H, Dean L, Collins R, Dodd R, Roberts A, Enghild J, Mantovani A, Fontana J, Baldock C, Inforzato A, Richter R, Day A
|
| RgGuinier |
8.6 |
nm |
| Dmax |
43.1 |
nm |
| VolumePorod |
658 |
nm3 |
|
|
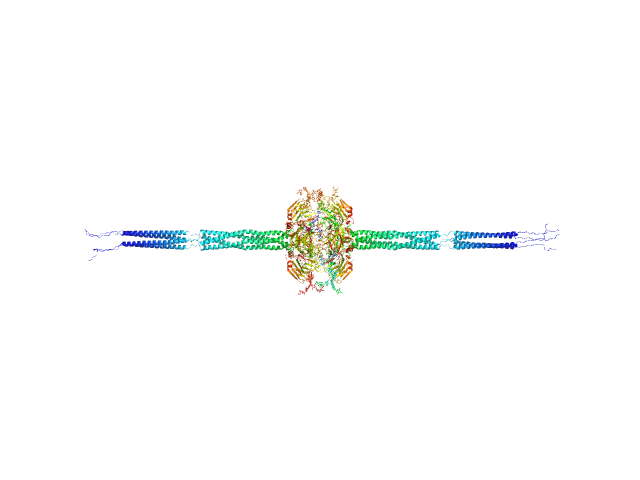
UniProt ID: P26022 (42-381) Pentraxin-related protein PTX3 (N-terminal deletion mutant)
|
|
|
|
| Sample: |
Pentraxin-related protein PTX3 (N-terminal deletion mutant) octamer, 301 kDa Homo sapiens protein
|
| Buffer: |
137 mM NaCl, 2.7 mM KCl, 10 mM phosphate buffer, pH: 7.4 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2020 Feb 6
|
The structural organisation of pentraxin-3 and its interactions with heavy chains of inter-α-inhibitor regulate crosslinking of the hyaluronan matrix
Matrix Biology 136:52-68 (2025)
Shah A, Zhang X, Snee M, Lockhart-Cairns M, Levy C, Jowitt T, Birchenough H, Dean L, Collins R, Dodd R, Roberts A, Enghild J, Mantovani A, Fontana J, Baldock C, Inforzato A, Richter R, Day A
|
| RgGuinier |
8.4 |
nm |
| Dmax |
40.0 |
nm |
| VolumePorod |
647 |
nm3 |
|
|
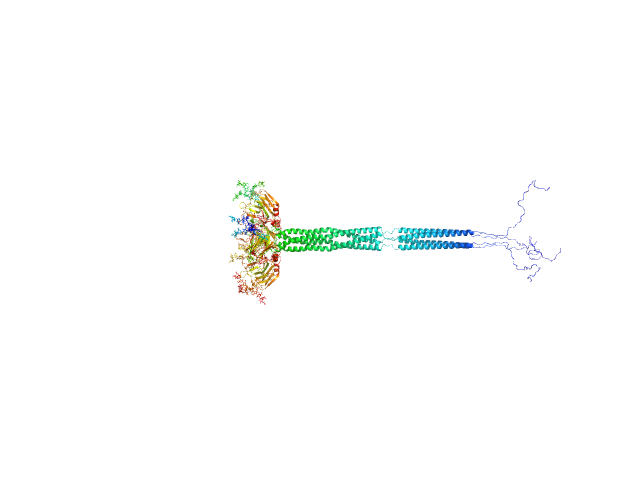
UniProt ID: P26022 (18-381) Pentraxin-related protein PTX3 (C317S, C318S)
|
|
|
|
| Sample: |
Pentraxin-related protein PTX3 (C317S, C318S) tetramer, 160 kDa Homo sapiens protein
|
| Buffer: |
137 mM NaCl, 2.7 mM KCl, 10 mM phosphate buffer, pH: 7.4 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2020 Feb 6
|
The structural organisation of pentraxin-3 and its interactions with heavy chains of inter-α-inhibitor regulate crosslinking of the hyaluronan matrix
Matrix Biology 136:52-68 (2025)
Shah A, Zhang X, Snee M, Lockhart-Cairns M, Levy C, Jowitt T, Birchenough H, Dean L, Collins R, Dodd R, Roberts A, Enghild J, Mantovani A, Fontana J, Baldock C, Inforzato A, Richter R, Day A
|
| RgGuinier |
7.8 |
nm |
| Dmax |
26.0 |
nm |
| VolumePorod |
370 |
nm3 |
|
|
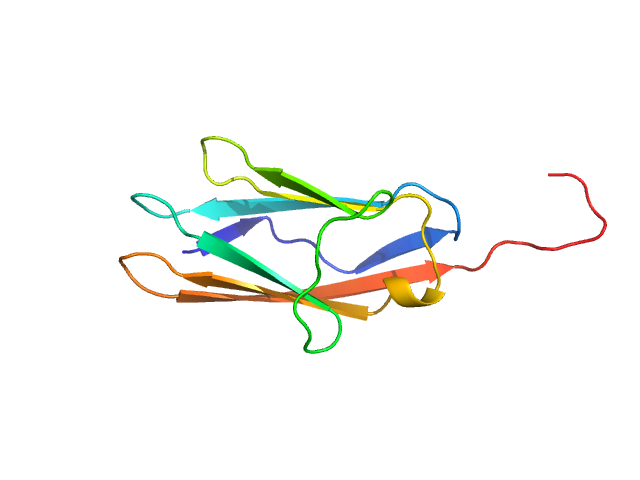
UniProt ID: Q9ET54 (1154-1257) Palladin
|
|
|
|
| Sample: |
Palladin monomer, 12 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES pH 7.4, 1 mM DTT, 100 mM NaCl, pH: |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2023 Aug 29
|
Integrated structural model of the palladin-actin complex using XL-MS, docking, NMR, and SAXS.
Protein Sci 34(5):e70122 (2025)
Sargent R, Liu DH, Yadav R, Glennenmeier D, Bradford C, Urbina N, Beck MR
|
| RgGuinier |
1.7 |
nm |
| Dmax |
6.8 |
nm |
| VolumePorod |
18 |
nm3 |
|
|
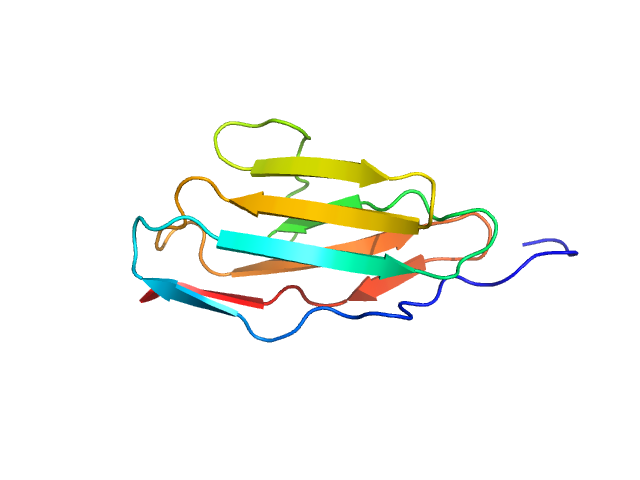
UniProt ID: Q9ET54 (1022-1126) Palladin
|
|
|
|
| Sample: |
Palladin monomer, 12 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES pH 7.4, 1 mM DTT, 100 mM NaCl, pH: |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2023 Aug 29
|
Integrated structural model of the palladin-actin complex using XL-MS, docking, NMR, and SAXS.
Protein Sci 34(5):e70122 (2025)
Sargent R, Liu DH, Yadav R, Glennenmeier D, Bradford C, Urbina N, Beck MR
|
| RgGuinier |
1.6 |
nm |
| Dmax |
6.7 |
nm |
| VolumePorod |
18 |
nm3 |
|
|
UniProt ID: Q9ET54 (1022-1257) Palladin
|
|
|
|
| Sample: |
Palladin monomer, 27 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES pH 7.4, 1 mM DTT, 100 mM NaCl, pH: |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2023 Aug 29
|
Integrated structural model of the palladin-actin complex using XL-MS, docking, NMR, and SAXS.
Protein Sci 34(5):e70122 (2025)
Sargent R, Liu DH, Yadav R, Glennenmeier D, Bradford C, Urbina N, Beck MR
|
| RgGuinier |
2.8 |
nm |
| Dmax |
12.3 |
nm |
| VolumePorod |
29 |
nm3 |
|
|
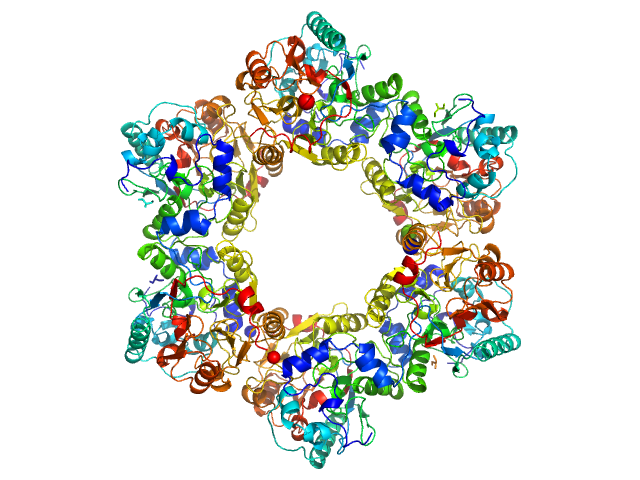
UniProt ID: F4ZCI3 (1-379) 2-nitroimidazole nitrohydrolase (T2I, G14D, K73R)
|
|
|
|
| Sample: |
2-nitroimidazole nitrohydrolase (T2I, G14D, K73R) hexamer, 258 kDa Mycobacterium sp. (strain … protein
|
| Buffer: |
25 mM Tris-HCl, 150 mM NaCl, 5% (v/v) glycerol, pH: 7.6 |
| Experiment: |
SAXS
data collected at BioSAXS, Australian Synchrotron on 2024 Aug 6
|
Structural insights into the enzymatic breakdown of azomycin-derived antibiotics by 2-nitroimdazole hydrolase (NnhA).
Commun Biol 7(1):1676 (2024)
Ahmed FH, Liu JW, Royan S, Warden AC, Esquirol L, Pandey G, Newman J, Scott C, Peat TS
|
| RgGuinier |
4.6 |
nm |
| Dmax |
12.6 |
nm |
| VolumePorod |
363 |
nm3 |
|
|
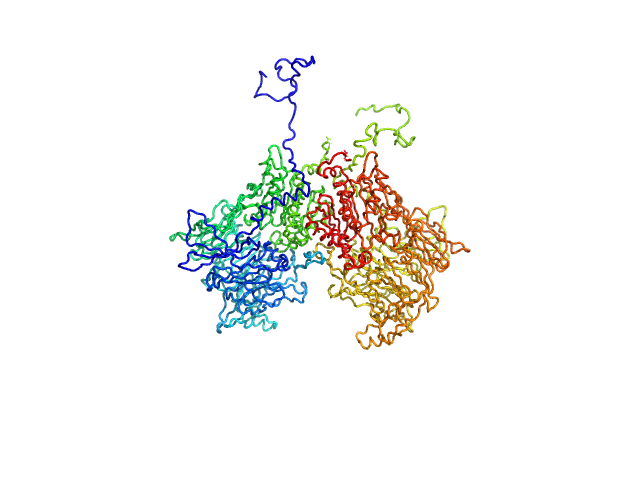
UniProt ID: Q6V1X1-1 (1-898) Isoform 1 of Dipeptidyl peptidase 8
|
|
|
|
| Sample: |
Isoform 1 of Dipeptidyl peptidase 8 dimer, 208 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, 2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2023 Oct 14
|
Computational study of DPP8 and DPP9: fundamental insights and inhibitor design
University of Antwerp PhD thesis c:irua:209673 (2024)
Olivier Beyens, Yann Sterckx
|
| RgGuinier |
4.2 |
nm |
| Dmax |
13.6 |
nm |
| VolumePorod |
271 |
nm3 |
|
|