UniProt ID: A9DF82 (2-117) Group 1 truncated hemoglobin (C51S, C71S, Y108A)
|
|
|
|
| Sample: |
Group 1 truncated hemoglobin (C51S, C71S, Y108A), 13 kDa Shewanella benthica KT99 protein
|
| Buffer: |
14 mM Tris, 6 mM potassium phosphate, pH: 7 |
| Experiment: |
SAXS
data collected at ID7A1 BioSAXS / HP-Bio Beamline, Cornell High Energy Synchrotron Source (CHESS) on 2023 Feb 18
|
Extremophilic hemoglobins: The structure of Shewanella benthica truncated hemoglobin N
Journal of Biological Chemistry :108223 (2025)
Martinez Grundman J, Schultz T, Schlessman J, Johnson E, Gillilan R, Lecomte J
|
| RgGuinier |
1.6 |
nm |
| Dmax |
6.0 |
nm |
| VolumePorod |
21 |
nm3 |
|
|
UniProt ID: A9DF82 (2-117) Group 1 truncated hemoglobin (C51S, C71S, Y108A)
|
|
|
|
| Sample: |
Group 1 truncated hemoglobin (C51S, C71S, Y108A), 13 kDa Shewanella benthica KT99 protein
|
| Buffer: |
14 mM Tris, 6 mM potassium phosphate, pH: 7 |
| Experiment: |
SAXS
data collected at ID7A1 BioSAXS / HP-Bio Beamline, Cornell High Energy Synchrotron Source (CHESS) on 2023 Feb 18
|
Extremophilic hemoglobins: The structure of Shewanella benthica truncated hemoglobin N
Journal of Biological Chemistry :108223 (2025)
Martinez Grundman J, Schultz T, Schlessman J, Johnson E, Gillilan R, Lecomte J
|
| RgGuinier |
1.8 |
nm |
| Dmax |
8.8 |
nm |
| VolumePorod |
24 |
nm3 |
|
|
UniProt ID: A9DF82 (2-117) Group 1 truncated hemoglobin (C51S, C71S, Y108A)
|
|
|
|
| Sample: |
Group 1 truncated hemoglobin (C51S, C71S, Y108A), 13 kDa Shewanella benthica KT99 protein
|
| Buffer: |
14 mM Tris, 6 mM potassium phosphate, pH: 7 |
| Experiment: |
SAXS
data collected at ID7A1 BioSAXS / HP-Bio Beamline, Cornell High Energy Synchrotron Source (CHESS) on 2023 Feb 18
|
Extremophilic hemoglobins: The structure of Shewanella benthica truncated hemoglobin N
Journal of Biological Chemistry :108223 (2025)
Martinez Grundman J, Schultz T, Schlessman J, Johnson E, Gillilan R, Lecomte J
|
| RgGuinier |
2.1 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
25 |
nm3 |
|
|
UniProt ID: A9DF82 (2-117) Group 1 truncated hemoglobin (C51S, C71S, Y108A)
|
|
|
|
| Sample: |
Group 1 truncated hemoglobin (C51S, C71S, Y108A), 13 kDa Shewanella benthica KT99 protein
|
| Buffer: |
14 mM Tris, 6 mM potassium phosphate, pH: 7 |
| Experiment: |
SAXS
data collected at ID7A1 BioSAXS / HP-Bio Beamline, Cornell High Energy Synchrotron Source (CHESS) on 2023 Feb 18
|
Extremophilic hemoglobins: The structure of Shewanella benthica truncated hemoglobin N
Journal of Biological Chemistry :108223 (2025)
Martinez Grundman J, Schultz T, Schlessman J, Johnson E, Gillilan R, Lecomte J
|
| RgGuinier |
2.2 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
26 |
nm3 |
|
|
UniProt ID: A9DF82 (2-117) Group 1 truncated hemoglobin (C51S, C71S, Y108A)
|
|
|
|
| Sample: |
Group 1 truncated hemoglobin (C51S, C71S, Y108A), 13 kDa Shewanella benthica KT99 protein
|
| Buffer: |
14 mM Tris, 6 mM potassium phosphate, pH: 7 |
| Experiment: |
SAXS
data collected at ID7A1 BioSAXS / HP-Bio Beamline, Cornell High Energy Synchrotron Source (CHESS) on 2023 Feb 18
|
Extremophilic hemoglobins: The structure of Shewanella benthica truncated hemoglobin N
Journal of Biological Chemistry :108223 (2025)
Martinez Grundman J, Schultz T, Schlessman J, Johnson E, Gillilan R, Lecomte J
|
| RgGuinier |
2.3 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
31 |
nm3 |
|
|
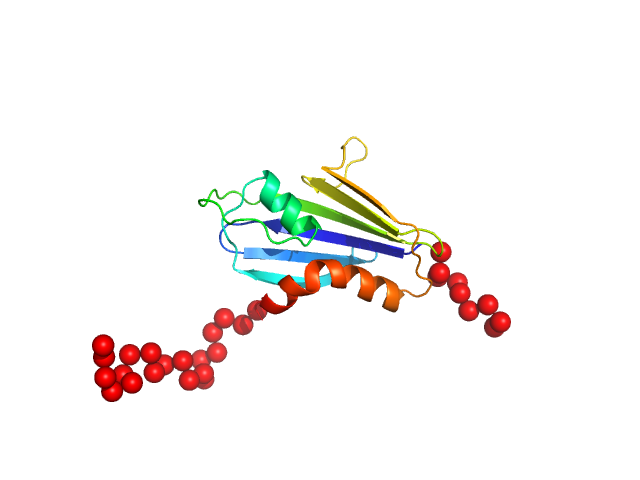
UniProt ID: Q88595 (923-1055) Genome polyprotein
|
|
|
|
| Sample: |
Genome polyprotein monomer, 17 kDa Theiler's murine encephalomyelitis … protein
|
| Buffer: |
20 mM HEPES pH 7.4, 1 M KCl, 2% v/v glycerol, 1 mM DTT, pH: |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Apr 26
|
A new protein-dependent riboswitch activates ribosomal frameshifting
(2025)
Betts J, Jeffries C, Passchier T, Kung H, Graham S, Abdelhamid M, Howard J, Craggs T, Graham S, Brierley I, Leake M, Quinn S, Hill C
|
| RgGuinier |
1.9 |
nm |
| Dmax |
7.2 |
nm |
| VolumePorod |
30 |
nm3 |
|
|
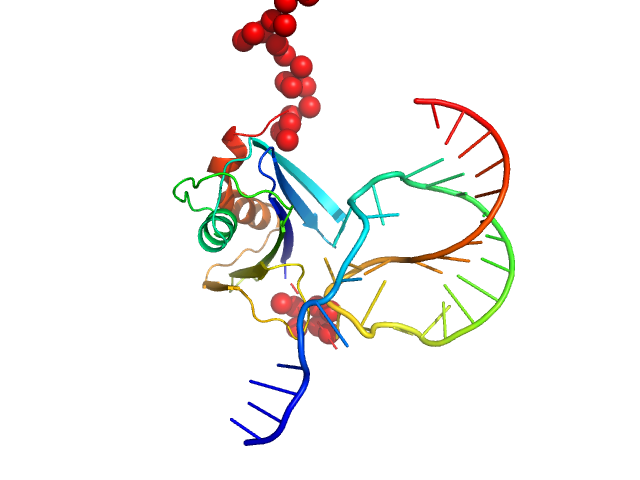
UniProt ID: Q88595 (923-1055) Genome polyprotein
UniProt ID: None (None-None) Stimulatory element RNA
|
|
|
|
| Sample: |
Genome polyprotein monomer, 17 kDa Theiler's murine encephalomyelitis … protein
Stimulatory element RNA monomer, 12 kDa synthetic RNA RNA
|
| Buffer: |
20 mM HEPES pH 7.4, 400 mM KCl 2% v/v glycerol, 1 mM DTT, pH: |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Apr 26
|
A new protein-dependent riboswitch activates ribosomal frameshifting
(2025)
Betts J, Jeffries C, Passchier T, Kung H, Graham S, Abdelhamid M, Howard J, Craggs T, Graham S, Brierley I, Leake M, Quinn S, Hill C
|
| RgGuinier |
2.2 |
nm |
| Dmax |
7.5 |
nm |
| VolumePorod |
46 |
nm3 |
|
|
UniProt ID: Q8DZX0 (311-412) ABC transporter, permease protein, extracellular domain
|
|
|
|
| Sample: |
ABC transporter, permease protein, extracellular domain monomer, 23 kDa Streptococcus agalactiae serotype … protein
|
| Buffer: |
25 mM MES, 500 mM NaCl, pH: 6 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2020 Dec 3
|
The extracellular domain of SaNSrFP binds bacitracin and allows the identification of new members of the BceAB transporter family
Jens Reiners
|
| RgGuinier |
2.4 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
50 |
nm3 |
|
|
UniProt ID: X5KGL2 (1-250) ABC transporter ATP-binding protein
UniProt ID: Q8DZX0 (1-651) ABC transporter, permease protein
|
|
|
|
| Sample: |
ABC transporter ATP-binding protein dimer, 62 kDa Streptococcus agalactiae protein
ABC transporter, permease protein monomer, 74 kDa Streptococcus agalactiae serotype … protein
|
| Buffer: |
50 mM Tris, 300 mM NaCl, 10 % Glycerol, 0.005 % LMNG,, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2023 Apr 25
|
The extracellular domain of SaNSrFP binds bacitracin and allows the identification of new members of the BceAB transporter family
Jens Reiners
|
| RgGuinier |
5.8 |
nm |
| Dmax |
20.5 |
nm |
| VolumePorod |
735 |
nm3 |
|
|
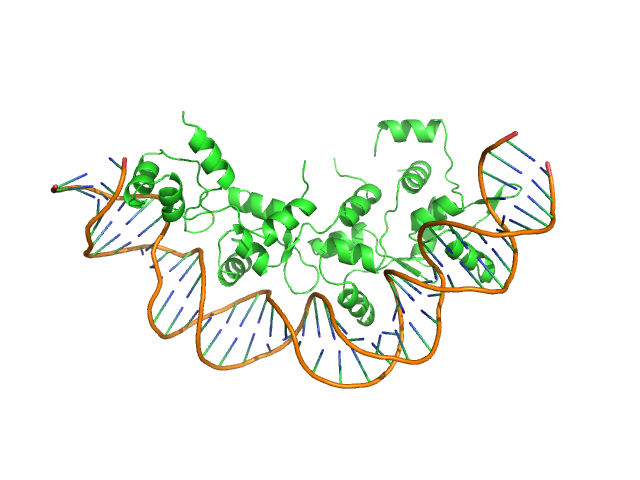
UniProt ID: Q7AL96 (None-None) Recombination directionality factor RdfS
UniProt ID: None (None-None) attP_8 40-mer DNA
|
|
|
|
| Sample: |
Recombination directionality factor RdfS tetramer, 52 kDa Mesorhizobium japonicum R7A protein
AttP_8 40-mer DNA dimer, 25 kDa DNA
|
| Buffer: |
150 mM Tris-HCl, 300 mM NaCl, 5% v/v glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2019 Jun 19
|
Structural basis for control of integrative and conjugative element excision and transfer by the oligomeric winged helix–turn–helix protein RdfS
Nucleic Acids Research 53(6) (2025)
Verdonk C, Agostino M, Eto K, Hall D, Bond C, Ramsay J
|
| RgGuinier |
3.8 |
nm |
| Dmax |
13.8 |
nm |
| VolumePorod |
134 |
nm3 |
|
|