UniProt ID: B5Z9C8 (1-117) Dihydroneopterin aldolase
|
|
|

|
| Sample: |
Dihydroneopterin aldolase tetramer, 55 kDa Helicobacter pylori (strain … protein
|
| Buffer: |
25 mM Tris-HCl pH 7.5 and 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2019 Apr 5
|
Structure of Helicobacter pylori dihydroneopterin aldolase suggests a fragment-based strategy for isozyme-specific inhibitor design.
Curr Res Struct Biol 5:100095 (2023)
Shaw GX, Fan L, Cherry S, Shi G, Tropea JE, Ji X
|
| RgGuinier |
2.5 |
nm |
| Dmax |
7.3 |
nm |
| VolumePorod |
77 |
nm3 |
|
|
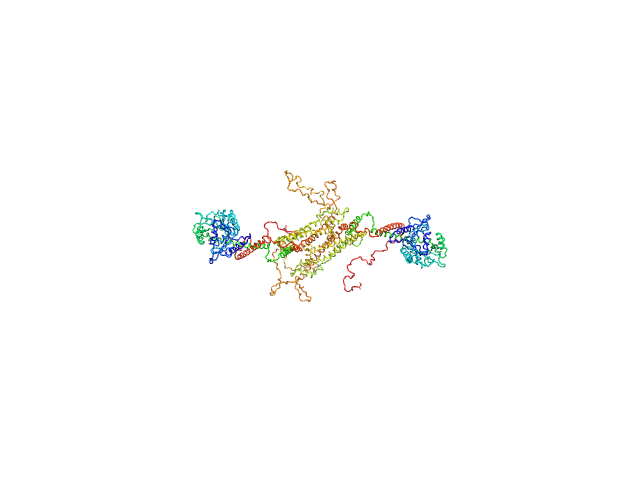
UniProt ID: E9BL28 (None-None) Dynamin central region family protein
|
|
|
|
| Sample: |
Dynamin central region family protein dimer, 165 kDa Leishmania donovani protein
|
| Buffer: |
20 mM HEPES, 200 mM NaCl, 3% glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2024 Apr 4
|
Biophysical analysis of an oligomerization-attenuated variant of the Leishmania donovani dynamin-1-like protein.
Mol Biochem Parasitol 263:111691 (2025)
Wuyts E, Sundaramoorthy R, Tulloch L, Monsieurs P, Eadsforth TC, Pintelon I, Timmermans JP, Dujardin JC, Domagalska MA, Caljon G, De Rycker M, Postis VLG, Wyllie S, Sterckx YG
|
| RgGuinier |
6.6 |
nm |
| Dmax |
25.0 |
nm |
| VolumePorod |
340 |
nm3 |
|
|
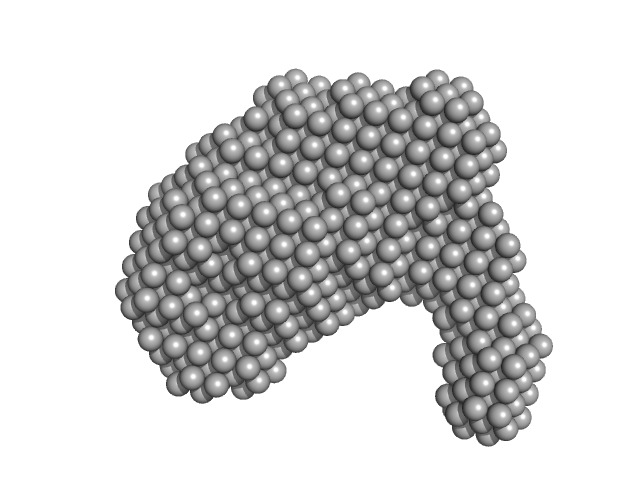
UniProt ID: O53943 (None-None) ESX-5 secretion-associated protein EspG5
UniProt ID: P9WPI1 (1-278) ESX-5 secretion system protein EccA5
|
|
|
|
| Sample: |
ESX-5 secretion-associated protein EspG5 monomer, 32 kDa Mycobacterium tuberculosis (strain … protein
ESX-5 secretion system protein EccA5 monomer, 31 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
50 mM HEPES, 200 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR-Central Drug Research Institute on 2025 Jan 28
|
Small angle x-ray scattering envelope of EccA5 N-terminal domain and EspG5 of Type 7 secretion pathway of Mycobacterium tuberculosis
Rajlakshmi K
|
| RgGuinier |
3.5 |
nm |
| Dmax |
9.1 |
nm |
| VolumePorod |
83 |
nm3 |
|
|
UniProt ID: P35755 (23-332) Iron-utilization periplasmic protein
|
|
|

|
| Sample: |
Iron-utilization periplasmic protein monomer, 34 kDa Haemophilus influenzae protein
|
| Buffer: |
1 mM Na2HPO4.7H2O, 0.18 mM KH2PO4, 13.7 mM NaCl, 0.27 mM KCl, 5%v/v Glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Aug 20
|
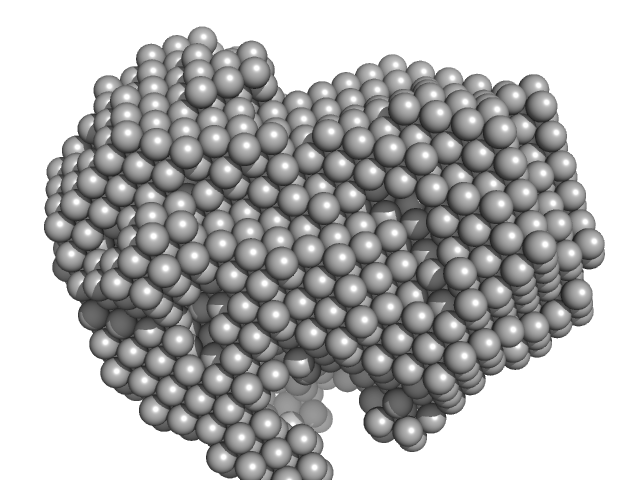
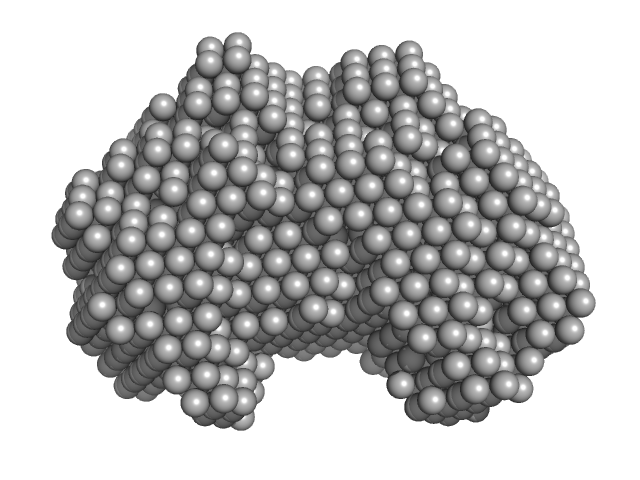
Conformational multiplicity of bacterial ferric binding protein revealed by small angle x-ray scattering and molecular dynamics calculations
The Journal of Chemical Physics 158(8) (2023)
Liu G, Ekmen E, Jalalypour F, Mertens H, Jeffries C, Svergun D, Atilgan A, Atilgan C, Sayers Z
|
| RgGuinier |
2.1 |
nm |
| Dmax |
6.3 |
nm |
| VolumePorod |
28 |
nm3 |
|
|
UniProt ID: P35755 (23-332) Iron-utilization periplasmic protein
|
|
|
|
| Sample: |
Iron-utilization periplasmic protein monomer, 34 kDa Haemophilus influenzae protein
|
| Buffer: |
10 mM Na2HPO4 . 7H2O 1.8 mM KH2PO4 137 mM NaCl 2.7 mM KCl 5% v/v Glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Aug 20
|
Conformational multiplicity of bacterial ferric binding protein revealed by small angle x-ray scattering and molecular dynamics calculations
The Journal of Chemical Physics 158(8) (2023)
Liu G, Ekmen E, Jalalypour F, Mertens H, Jeffries C, Svergun D, Atilgan A, Atilgan C, Sayers Z
|
| RgGuinier |
2.0 |
nm |
| Dmax |
6.2 |
nm |
| VolumePorod |
34 |
nm3 |
|
|
UniProt ID: P35755 (23-332) Iron-utilization periplasmic protein
|
|
|
|
| Sample: |
Iron-utilization periplasmic protein monomer, 34 kDa Haemophilus influenzae protein
|
| Buffer: |
10 mM Na2HPO4 . 7H2O 1.8 mM KH2PO4 137 mM NaCl 2.7 mM KCl 5% v/v Glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Dec 9
|
Conformational multiplicity of bacterial ferric binding protein revealed by small angle x-ray scattering and molecular dynamics calculations
The Journal of Chemical Physics 158(8) (2023)
Liu G, Ekmen E, Jalalypour F, Mertens H, Jeffries C, Svergun D, Atilgan A, Atilgan C, Sayers Z
|
| RgGuinier |
2.1 |
nm |
| Dmax |
6.5 |
nm |
| VolumePorod |
39 |
nm3 |
|
|
UniProt ID: P35755 (23-332) Iron-utilization periplasmic protein
|
|
|

|
| Sample: |
Iron-utilization periplasmic protein monomer, 34 kDa Haemophilus influenzae protein
|
| Buffer: |
10 mM Na2HPO4 . 7H2O 1.8 mM KH2PO4 137 mM NaCl 2.7 mM KCl 5% v/v Glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Dec 9
|
Conformational multiplicity of bacterial ferric binding protein revealed by small angle x-ray scattering and molecular dynamics calculations
The Journal of Chemical Physics 158(8) (2023)
Liu G, Ekmen E, Jalalypour F, Mertens H, Jeffries C, Svergun D, Atilgan A, Atilgan C, Sayers Z
|
| RgGuinier |
2.0 |
nm |
| Dmax |
5.9 |
nm |
| VolumePorod |
33 |
nm3 |
|
|
UniProt ID: P35755 (23-332) Iron-utilization periplasmic protein
|
|
|

|
| Sample: |
Iron-utilization periplasmic protein monomer, 34 kDa Haemophilus influenzae protein
|
| Buffer: |
10 mM Na2HPO4 . 7H2O 1.8 mM KH2PO4 137 mM NaCl 2.7 mM KCl 5% v/v Glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Sep 19
|
Conformational multiplicity of bacterial ferric binding protein revealed by small angle x-ray scattering and molecular dynamics calculations
The Journal of Chemical Physics 158(8) (2023)
Liu G, Ekmen E, Jalalypour F, Mertens H, Jeffries C, Svergun D, Atilgan A, Atilgan C, Sayers Z
|
| RgGuinier |
2.1 |
nm |
| Dmax |
6.2 |
nm |
| VolumePorod |
40 |
nm3 |
|
|
UniProt ID: P35755 (23-332) Iron-utilization periplasmic protein
|
|
|

|
| Sample: |
Iron-utilization periplasmic protein monomer, 34 kDa Haemophilus influenzae protein
|
| Buffer: |
1 mM Na2HPO4.7H2O, 0.18 mM KH2PO4, 13.7 mM NaCl, 0.27 mM KCl, 5%v/v Glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Aug 20
|
Conformational multiplicity of bacterial ferric binding protein revealed by small angle x-ray scattering and molecular dynamics calculations
The Journal of Chemical Physics 158(8) (2023)
Liu G, Ekmen E, Jalalypour F, Mertens H, Jeffries C, Svergun D, Atilgan A, Atilgan C, Sayers Z
|
| RgGuinier |
2.0 |
nm |
| Dmax |
6.1 |
nm |
| VolumePorod |
24 |
nm3 |
|
|
UniProt ID: P35755 (23-332) Iron-utilization periplasmic protein
|
|
|
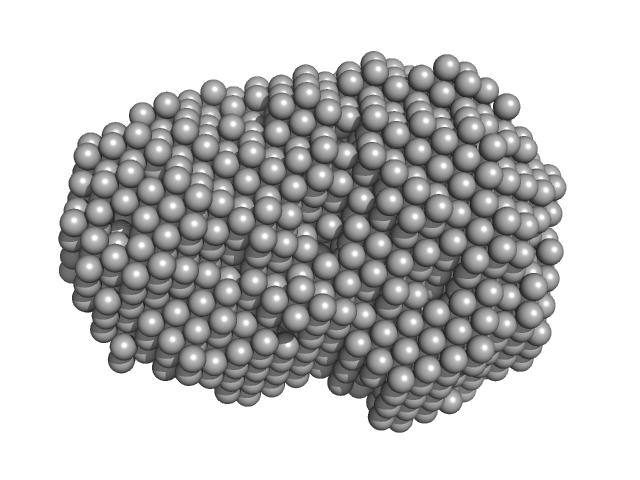
|
| Sample: |
Iron-utilization periplasmic protein monomer, 34 kDa Haemophilus influenzae protein
|
| Buffer: |
1 mM Na2HPO4.7H2O, 0.18 mM KH2PO4, 13.7 mM NaCl, 0.27 mM KCl, 5%v/v Glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Aug 20
|
Conformational multiplicity of bacterial ferric binding protein revealed by small angle x-ray scattering and molecular dynamics calculations
The Journal of Chemical Physics 158(8) (2023)
Liu G, Ekmen E, Jalalypour F, Mertens H, Jeffries C, Svergun D, Atilgan A, Atilgan C, Sayers Z
|
| RgGuinier |
2.1 |
nm |
| Dmax |
6.3 |
nm |
| VolumePorod |
35 |
nm3 |
|
|