UniProt ID: P12004 (1-261) Proliferating cell nuclear antigen
UniProt ID: Q15004 (2-111) PCNA-associated factor
|
|
|
|
| Sample: |
Proliferating cell nuclear antigen trimer, 87 kDa Homo sapiens protein
PCNA-associated factor monomer, 12 kDa Homo sapiens protein
|
| Buffer: |
137 mM NaCl, 2.7 mM KCl, 10 mM sodium phosphate, 2 mM potassium phosphate (PBS), pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2012 Apr 23
|
KDSAXS: A tool for Analyzing Binding Equilibria with SAXS Data using Explicit Models
Journal of Molecular Biology :169103 (2025)
Gomes T, Ruiz L, Martin-Malpartida P, Bernadó P, Baptista A, Macias M, Cordeiro T
|
| RgGuinier |
3.5 |
nm |
| Dmax |
9.7 |
nm |
| VolumePorod |
128 |
nm3 |
|
|
UniProt ID: P12004 (1-261) Proliferating cell nuclear antigen
UniProt ID: Q15004 (2-111) PCNA-associated factor
|
|
|
|
| Sample: |
Proliferating cell nuclear antigen trimer, 87 kDa Homo sapiens protein
PCNA-associated factor monomer, 12 kDa Homo sapiens protein
|
| Buffer: |
137 mM NaCl, 2.7 mM KCl, 10 mM sodium phosphate, 2 mM potassium phosphate (PBS), pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2012 Apr 23
|
KDSAXS: A tool for Analyzing Binding Equilibria with SAXS Data using Explicit Models
Journal of Molecular Biology :169103 (2025)
Gomes T, Ruiz L, Martin-Malpartida P, Bernadó P, Baptista A, Macias M, Cordeiro T
|
| RgGuinier |
3.5 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
158 |
nm3 |
|
|
UniProt ID: P12004 (1-261) Proliferating cell nuclear antigen
UniProt ID: Q15004 (2-111) PCNA-associated factor
|
|
|
|
| Sample: |
Proliferating cell nuclear antigen trimer, 87 kDa Homo sapiens protein
PCNA-associated factor monomer, 12 kDa Homo sapiens protein
|
| Buffer: |
137 mM NaCl, 2.7 mM KCl, 10 mM sodium phosphate, 2 mM potassium phosphate (PBS), pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2012 Apr 23
|
KDSAXS: A tool for Analyzing Binding Equilibria with SAXS Data using Explicit Models
Journal of Molecular Biology :169103 (2025)
Gomes T, Ruiz L, Martin-Malpartida P, Bernadó P, Baptista A, Macias M, Cordeiro T
|
| RgGuinier |
3.7 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
184 |
nm3 |
|
|
UniProt ID: P12004 (1-261) Proliferating cell nuclear antigen
UniProt ID: Q15004 (2-111) PCNA-associated factor
|
|
|
|
| Sample: |
Proliferating cell nuclear antigen trimer, 87 kDa Homo sapiens protein
PCNA-associated factor monomer, 12 kDa Homo sapiens protein
|
| Buffer: |
137 mM NaCl, 2.7 mM KCl, 10 mM sodium phosphate, 2 mM potassium phosphate (PBS), pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2012 Apr 23
|
KDSAXS: A tool for Analyzing Binding Equilibria with SAXS Data using Explicit Models
Journal of Molecular Biology :169103 (2025)
Gomes T, Ruiz L, Martin-Malpartida P, Bernadó P, Baptista A, Macias M, Cordeiro T
|
| RgGuinier |
3.7 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
219 |
nm3 |
|
|
UniProt ID: P12004 (1-261) Proliferating cell nuclear antigen
UniProt ID: Q15004 (2-111) PCNA-associated factor
|
|
|
|
| Sample: |
Proliferating cell nuclear antigen trimer, 87 kDa Homo sapiens protein
PCNA-associated factor monomer, 12 kDa Homo sapiens protein
|
| Buffer: |
137 mM NaCl, 2.7 mM KCl, 10 mM sodium phosphate, 2 mM potassium phosphate (PBS), pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2012 Apr 23
|
KDSAXS: A tool for Analyzing Binding Equilibria with SAXS Data using Explicit Models
Journal of Molecular Biology :169103 (2025)
Gomes T, Ruiz L, Martin-Malpartida P, Bernadó P, Baptista A, Macias M, Cordeiro T
|
| RgGuinier |
3.7 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
208 |
nm3 |
|
|
UniProt ID: P12004 (1-261) Proliferating cell nuclear antigen
UniProt ID: Q15004 (2-111) PCNA-associated factor
|
|
|
|
| Sample: |
Proliferating cell nuclear antigen trimer, 87 kDa Homo sapiens protein
PCNA-associated factor monomer, 12 kDa Homo sapiens protein
|
| Buffer: |
137 mM NaCl, 2.7 mM KCl, 10 mM sodium phosphate, 2 mM potassium phosphate (PBS), pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2012 Apr 23
|
KDSAXS: A tool for Analyzing Binding Equilibria with SAXS Data using Explicit Models
Journal of Molecular Biology :169103 (2025)
Gomes T, Ruiz L, Martin-Malpartida P, Bernadó P, Baptista A, Macias M, Cordeiro T
|
| RgGuinier |
3.9 |
nm |
| Dmax |
11.5 |
nm |
| VolumePorod |
194 |
nm3 |
|
|
UniProt ID: P12004 (1-261) Proliferating cell nuclear antigen
UniProt ID: Q15004 (2-111) PCNA-associated factor
|
|
|
|
| Sample: |
Proliferating cell nuclear antigen trimer, 87 kDa Homo sapiens protein
PCNA-associated factor monomer, 12 kDa Homo sapiens protein
|
| Buffer: |
137 mM NaCl, 2.7 mM KCl, 10 mM sodium phosphate, 2 mM potassium phosphate (PBS), pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2012 Apr 23
|
KDSAXS: A tool for Analyzing Binding Equilibria with SAXS Data using Explicit Models
Journal of Molecular Biology :169103 (2025)
Gomes T, Ruiz L, Martin-Malpartida P, Bernadó P, Baptista A, Macias M, Cordeiro T
|
| RgGuinier |
3.7 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
187 |
nm3 |
|
|
UniProt ID: Q92997 (245-351) Segment polarity protein dishevelled homolog DVL-3
|
|
|
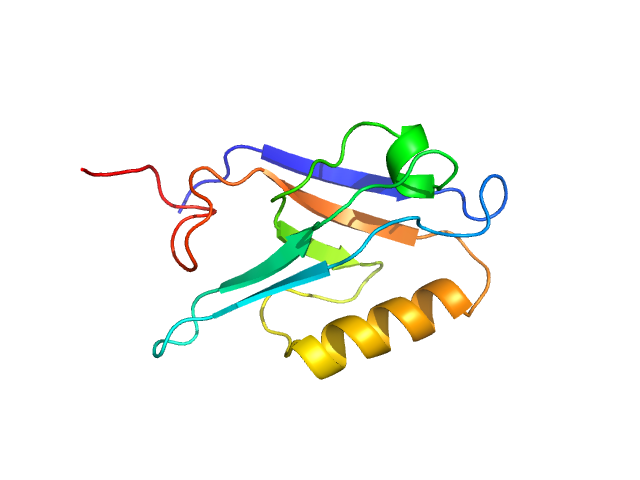
|
| Sample: |
Segment polarity protein dishevelled homolog DVL-3 monomer, 12 kDa Homo sapiens protein
|
| Buffer: |
50 mM sodium phosphate, 50 mM NaCl, 0.5 mM EDTA, pH: 6.5 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-2000, CEITEC on 2021 Jan 27
|
A class III ligand oscillates between internal and terminal binding modes as it engages with the Dishevelled PDZ domain.
Structure (2025)
Kumar J, Micka M, Komárek J, Klumpler T, Bystrý V, Sprangers R, Bařinka C, Bryja V, Tripsianes K
|
| RgGuinier |
1.6 |
nm |
| Dmax |
4.3 |
nm |
| VolumePorod |
20 |
nm3 |
|
|
UniProt ID: A8JD56 (68-548) peptidylprolyl isomerase
|
|
|

|
| Sample: |
Peptidylprolyl isomerase monomer, 54 kDa Chlamydomonas reinhardtii protein
|
| Buffer: |
20 mM Tris pH 7.5, 150 mM KCl, pH: |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Apr 30
|
Structural and molecular comparison of bacterial and eukaryotic trigger factors.
Sci Rep 7(1):10680 (2017)
Ries F, Carius Y, Rohr M, Gries K, Keller S, Lancaster CRD, Willmund F
|
| RgGuinier |
3.9 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
103 |
nm3 |
|
|
UniProt ID: Q8S9L5 (78-547) Trigger factor-like protein TIG, Chloroplastic
|
|
|

|
| Sample: |
Trigger factor-like protein TIG, Chloroplastic monomer, 53 kDa Arabidopsis thaliana protein
|
| Buffer: |
20 mM Tris pH 7.5, 150 mM KCl, pH: |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Apr 30
|
Structural and molecular comparison of bacterial and eukaryotic trigger factors.
Sci Rep 7(1):10680 (2017)
Ries F, Carius Y, Rohr M, Gries K, Keller S, Lancaster CRD, Willmund F
|
| RgGuinier |
3.8 |
nm |
| Dmax |
12.7 |
nm |
| VolumePorod |
104 |
nm3 |
|
|