UniProt ID: Q9AES2 (31-1166) IgA protease (E540A)
|
|
|
|
| Sample: |
IgA protease (E540A) monomer, 128 kDa Thomasclavelia ramosa protein
|
| Buffer: |
50 mM Tris, 150 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2023 May 23
|
Biochemical and structural characterization of the human gut microbiome metallopeptidase IgAse provides insight into its unique specificity for the Fab' region of IgA1 and IgA2.
PLoS Pathog 21(7):e1013292 (2025)
Ramírez-Larrota JS, Juyoux P, Guerra P, Eckhard U, Gomis-Rüth FX
|
| RgGuinier |
5.7 |
nm |
| Dmax |
27.9 |
nm |
| VolumePorod |
190 |
nm3 |
|
|
UniProt ID: O07411 (1-554) Probable fatty-acid-CoA ligase FadD5 (Fatty-acid-CoA synthetase) (Fatty-acid-CoA synthase)
|
|
|
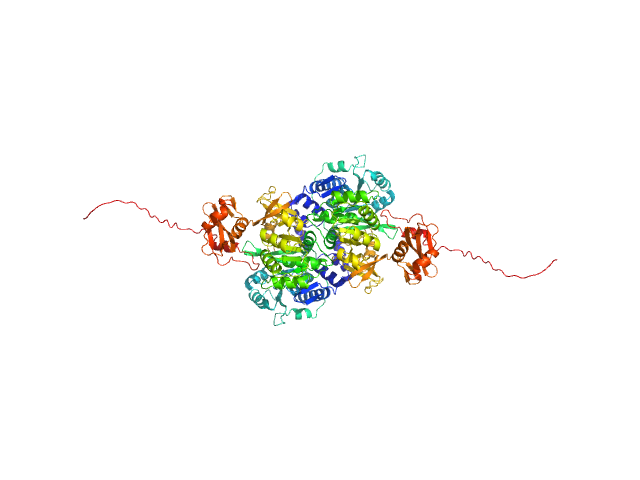
|
| Sample: |
Probable fatty-acid-CoA ligase FadD5 (Fatty-acid-CoA synthetase) (Fatty-acid-CoA synthase) dimer, 123 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
20 mM HEPES, 500 mM NaCl, 5 mM MgCl2, 1 mM β-mercaptoethanol, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 Oct 20
|
Structural enzymological studies of the long chain fatty acyl-CoA synthetase FadD5 from the mce1 operon of Mycobacterium tuberculosis.
Biochem Biophys Res Commun 769:151960 (2025)
Rahman MA, Dalwani S, Venkatesan R
|
| RgGuinier |
3.6 |
nm |
| Dmax |
12.3 |
nm |
| VolumePorod |
221 |
nm3 |
|
|
UniProt ID: O07411 (1-554) Probable fatty-acid-CoA ligase FadD5 (Fatty-acid-CoA synthetase) (Fatty-acid-CoA synthase)
|
|
|
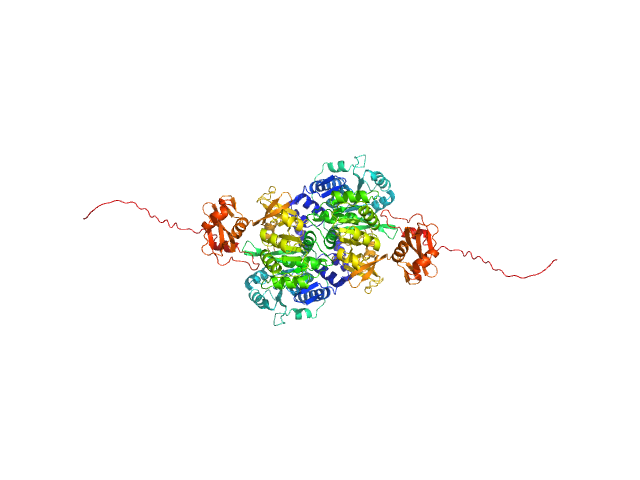
|
| Sample: |
Probable fatty-acid-CoA ligase FadD5 (Fatty-acid-CoA synthetase) (Fatty-acid-CoA synthase) dimer, 123 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
20 mM HEPES, 500 mM NaCl, 5 mM MgCl2, 1 mM β-mercaptoethanol, 2mM ATP, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 Oct 20
|
Structural enzymological studies of the long chain fatty acyl-CoA synthetase FadD5 from the mce1 operon of Mycobacterium tuberculosis.
Biochem Biophys Res Commun 769:151960 (2025)
Rahman MA, Dalwani S, Venkatesan R
|
| RgGuinier |
3.6 |
nm |
| Dmax |
12.1 |
nm |
| VolumePorod |
213 |
nm3 |
|
|
UniProt ID: O07411 (1-554) Probable fatty-acid-CoA ligase FadD5 (Fatty-acid-CoA synthetase) (Fatty-acid-CoA synthase)
|
|
|

|
| Sample: |
Probable fatty-acid-CoA ligase FadD5 (Fatty-acid-CoA synthetase) (Fatty-acid-CoA synthase) dimer, 123 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
20 mM HEPES, 500 mM NaCl, 5 mM MgCl2, 1 mM β-mercaptoethanol, 2 mM coenzyme A (CoA), pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 Oct 20
|
Structural enzymological studies of the long chain fatty acyl-CoA synthetase FadD5 from the mce1 operon of Mycobacterium tuberculosis.
Biochem Biophys Res Commun 769:151960 (2025)
Rahman MA, Dalwani S, Venkatesan R
|
| RgGuinier |
3.7 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
210 |
nm3 |
|
|
UniProt ID: O07419 (107-213) Probable conserved Mce associated membrane protein
|
|
|
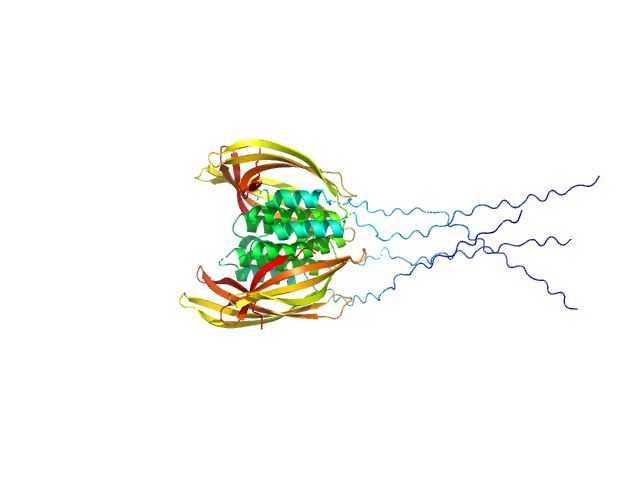
|
| Sample: |
Probable conserved Mce associated membrane protein tetramer, 59 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
50mM Tris, 500mM NaCl, pH: 8.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Dec 10
|
Insight into the structure and interactions of the M. tuberculosis
Mce-associated membrane proteins Mam1A-1D
(2025)
Hynönen M, Perumal P, Hynönen N, Doutch J, Ma K, Venkatesan R
|
| RgGuinier |
3.2 |
nm |
| Dmax |
90.0 |
nm |
| VolumePorod |
90 |
nm3 |
|
|
UniProt ID: O07419 (107-213) Probable conserved Mce associated membrane protein
|
|
|

|
| Sample: |
Probable conserved Mce associated membrane protein tetramer, 59 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
50 mM Tris, 500 mM NaCl, D-C12E9, pH: 8.5 |
| Experiment: |
SANS
data collected at SANS2D, ISIS Neutron and Muon Source on 2019 Dec 10
|
Insight into the structure and interactions of the M. tuberculosis
Mce-associated membrane proteins Mam1A-1D
(2025)
Hynönen M, Perumal P, Hynönen N, Doutch J, Ma K, Venkatesan R
|
| RgGuinier |
2.6 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
33 |
nm3 |
|
|
UniProt ID: B8BQS1 (None-None) CP12 domain-containing protein
|
|
|

|
| Sample: |
CP12 domain-containing protein dimer, 34 kDa Thalassiosira pseudonana protein
|
| Buffer: |
30 mM Tris, 50 mM NaCl, 2 mM EDTA, 1 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Feb 23
|
A new type of flexible CP12 protein in the marine diatom Thalassiosira pseudonana
Cell Communication and Signaling 19(1) (2021)
Shao H, Huang W, Avilan L, Receveur-Bréchot V, Puppo C, Puppo R, Lebrun R, Gontero B, Launay H
|
| RgGuinier |
3.8 |
nm |
| Dmax |
13.5 |
nm |
|
|
UniProt ID: Q93IQ4 (151-509) Alkaline serine protease
|
|
|
|
| Sample: |
Alkaline serine protease monomer, 36 kDa Stenotrophomonas maltophilia protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2023 Jun 10
|
Unveiling the structure, function and dynamics of StmPr1 in Stenotrophomonas maltophilia virulence.
Sci Rep 15(1):20193 (2025)
Sommer M, Negm A, Outzen L, Windhorst S, Gabdulkhakov A, Weber W, Betzel C
|
| RgGuinier |
1.9 |
nm |
| Dmax |
5.5 |
nm |
| VolumePorod |
44 |
nm3 |
|
|
UniProt ID: Q93IQ4 (151-617) Alkaline serine protease (Δ1-150)
|
|
|
|
| Sample: |
Alkaline serine protease (Δ1-150) monomer, 47 kDa Stenotrophomonas maltophilia protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Nov 25
|
Unveiling the structure, function and dynamics of StmPr1 in Stenotrophomonas maltophilia virulence.
Sci Rep 15(1):20193 (2025)
Sommer M, Negm A, Outzen L, Windhorst S, Gabdulkhakov A, Weber W, Betzel C
|
| RgGuinier |
2.9 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
112 |
nm3 |
|
|
UniProt ID: P02766 (21-147) Transthyretin
|
|
|
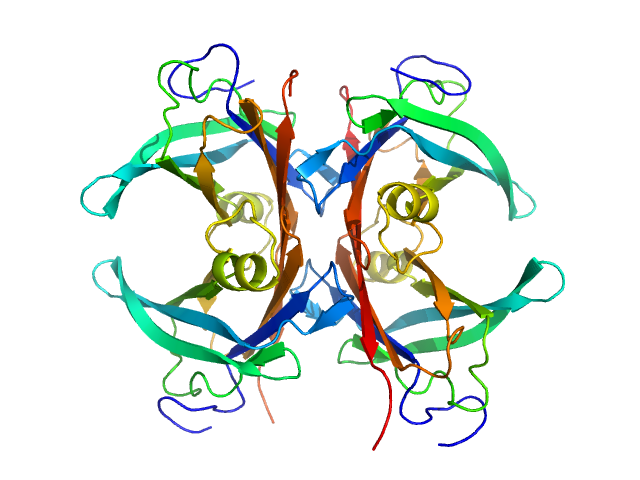
|
| Sample: |
Transthyretin tetramer, 55 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 50 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at 13A, Taiwan Photon Source, NSRRC on 2022 Sep 4
|
Differentiating the solution structures and stability of transthyretin tetramer complexed with tolcapone and tafamidis using SEC-SWAXS and NMR.
J Appl Crystallogr 58(Pt 4):1373-1383 (2025)
Shih O, Feng YC, Agrawal S, Liao KF, Yeh YQ, Chang JW, Yu TY, Jeng US
|
| RgGuinier |
2.4 |
nm |
| Dmax |
7.2 |
nm |
| VolumePorod |
92 |
nm3 |
|
|