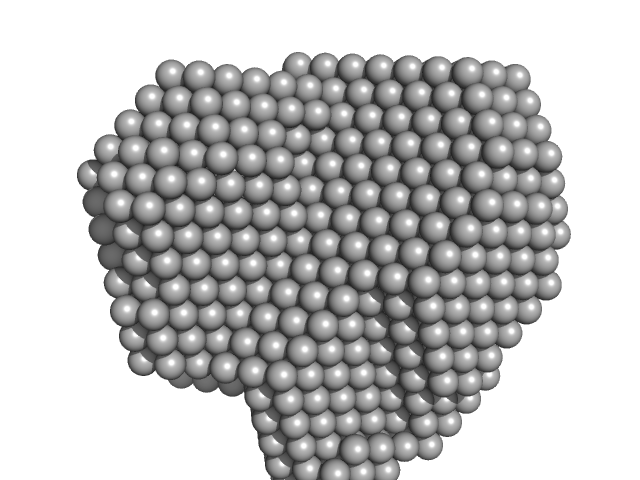
UniProt ID: P9WNV1 (601-691) M.tb. LigA BRCT domain (DNA ligase A)
|
|
|
|
| Sample: |
M.tb. LigA BRCT domain (DNA ligase A) monomer, 13 kDa Mycobacterium tuberculosis protein
|
| Buffer: |
50 mM Tris-HCl 500 mM NaCl 5mM β-mercaptoethanol, pH: 8 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR-Central Drug Research Institute on 2018 Jun 2
|
M. tuberculosis class II apurinic/ apyrimidinic-endonuclease/3'-5' exonuclease (XthA) engages with NAD+-dependent DNA ligase A (LigA) to counter futile cleavage and ligation cycles in base excision repair.
Nucleic Acids Res (2020)
Khanam T, Afsar M, Shukla A, Alam F, Kumar S, Soyar H, Dolma K, Pasupuleti M, Srivastava KK, Ampapathi RS, Ramachandran R
|
| RgGuinier |
1.6 |
nm |
| Dmax |
3.7 |
nm |
| VolumePorod |
23 |
nm3 |
|
|
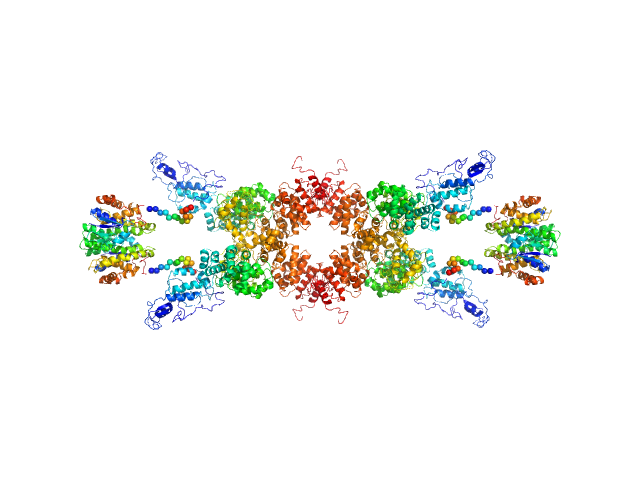
UniProt ID: Q9LM76 (None-None) Senescence-associated E3 ubiquitin ligase 1
UniProt ID: P08515 (None-None) Glutathione S-transferase
|
|
|
|
| Sample: |
Senescence-associated E3 ubiquitin ligase 1 tetramer, 355 kDa Arabidopsis thaliana protein
Glutathione S-transferase tetramer, 106 kDa Schistosoma japonicum protein
|
| Buffer: |
50 mM Tris, 250 mM NaCl, pH: 9 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2014 Jan 20
|
Senescence-associated ubiquitin ligase 1 (SAUL1)
Haifa El Kilani,
Al Kikhney
|
| RgGuinier |
7.6 |
nm |
| Dmax |
28.0 |
nm |
| VolumePorod |
778 |
nm3 |
|
|
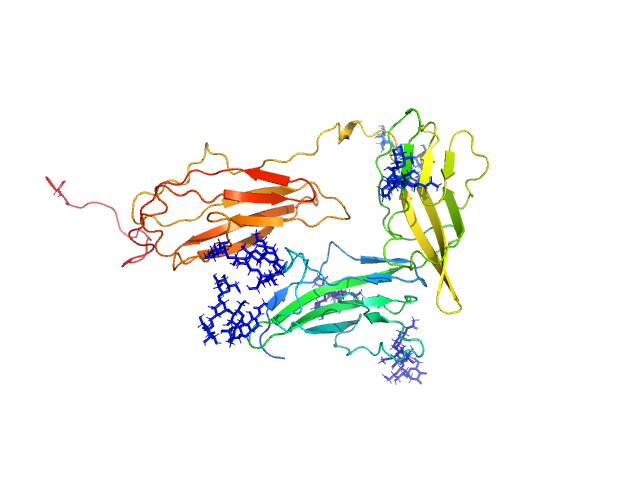
UniProt ID: Q9NPH3 (21-348) Interleukin-1 receptor accessory protein ectodomains with ST2 linker
|
|
|
|
| Sample: |
Interleukin-1 receptor accessory protein ectodomains with ST2 linker monomer, 41 kDa Homo sapiens protein
|
| Buffer: |
10mM HEPES, 150mM NaCl, 3% glycerol, pH: 7.2 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2017 Jul 24
|
Functional Relevance of Interleukin-1 Receptor Inter-domain Flexibility for Cytokine Binding and Signaling.
Structure 27(8):1296-1307.e5 (2019)
Ge J, Remesh SG, Hammel M, Pan S, Mahan AD, Wang S, Wang X
|
| RgGuinier |
3.1 |
nm |
| Dmax |
10.7 |
nm |
| VolumePorod |
76 |
nm3 |
|
|
UniProt ID: None (None-None) 80bp_DNA Forward
UniProt ID: None (None-None) 80bp_DNA Reverse
UniProt ID: P0ACF0 (1-90) DNA-binding protein HU-alpha
|
|
|
|
| Sample: |
80bp_DNA Forward monomer, 25 kDa Escherichia coli DNA
80bp_DNA Reverse monomer, 25 kDa Escherichia coli DNA
DNA-binding protein HU-alpha, 10 kDa Escherichia coli protein
|
| Buffer: |
10 mM sodium acetate, 50 mM NaCl, pH: 4.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 May 27
|
Nucleoid remodeling during environmental adaptation is regulated by HU-dependent DNA bundling.
Nat Commun 11(1):2905 (2020)
Remesh SG, Verma SC, Chen JH, Ekman AA, Larabell CA, Adhya S, Hammel M
|
|
|
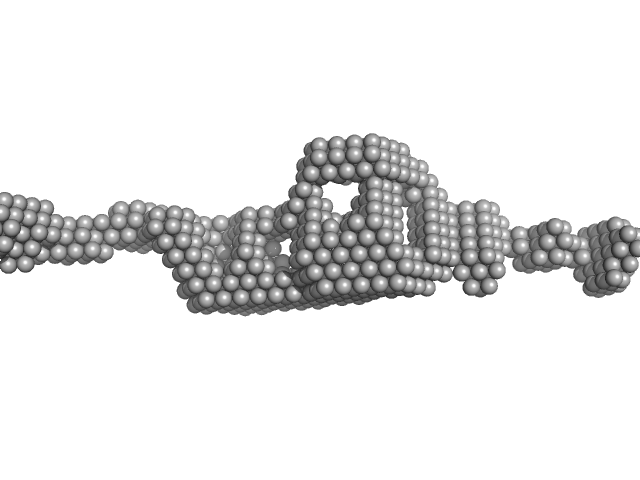
UniProt ID: P63034 (2-400) Cytohesin-2
|
|
|
|
| Sample: |
Cytohesin-2 dimer, 95 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 2 mM MgCl2, 0.1% 2-mercaptoethanol, 5% glycerol, 0.001 mM insitol 1,3,4,5-tetrakis phosphate, pH: 8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2013 Nov 15
|
Structural Organization and Dynamics of Homodimeric Cytohesin Family Arf GTPase Exchange Factors in Solution and on Membranes.
Structure (2019)
Das S, Malaby AW, Nawrotek A, Zhang W, Zeghouf M, Maslen S, Skehel M, Chakravarthy S, Irving TC, Bilsel O, Cherfils J, Lambright DG
|
| RgGuinier |
5.3 |
nm |
| Dmax |
27.0 |
nm |
| VolumePorod |
180 |
nm3 |
|
|
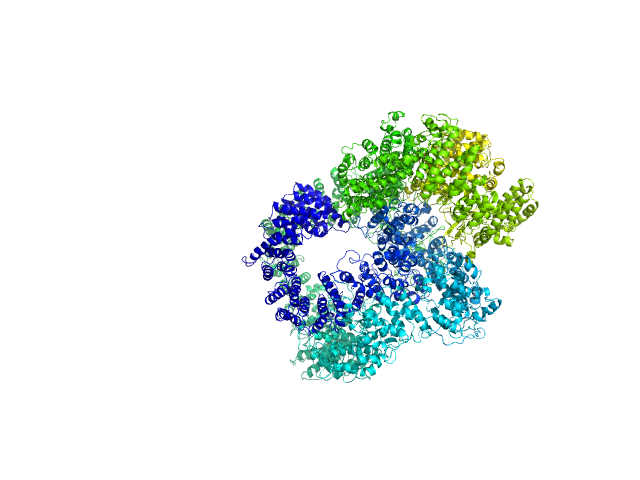
UniProt ID: P12956 (1-609) X-ray repair cross-complementing protein 6
UniProt ID: P13010 (1-732) X-ray repair cross-complementing protein 5
UniProt ID: P78527 (10-4128) DNA-dependent protein kinase catalytic subunit
UniProt ID: None (None-None) dsDNA
|
|
|
|
| Sample: |
X-ray repair cross-complementing protein 6 monomer, 70 kDa Homo sapiens protein
X-ray repair cross-complementing protein 5 monomer, 83 kDa Homo sapiens protein
DNA-dependent protein kinase catalytic subunit monomer, 468 kDa Homo sapiens protein
DsDNA dimer, 21 kDa DNA
|
| Buffer: |
50 mM Tris-HCl, 100 mM NaCl, 5% glycerol, 0.01% sodium azide, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2016 Dec 30
|
Visualizing functional dynamicity in the DNA-dependent protein kinase holoenzyme DNA-PK complex by integrating SAXS with cryo-EM.
Prog Biophys Mol Biol (2020)
Hammel M, Rosenberg DJ, Bierma J, Hura GL, Lees-Miller SP, Tainer JA
|
| RgGuinier |
7.5 |
nm |
| Dmax |
29.4 |
nm |
| VolumePorod |
1440 |
nm3 |
|
|
UniProt ID: None (None-None) n-Dodecyl-β-D-Maltopyranoside
UniProt ID: I6YC99 (121-400) Mce-family protein Mce4A
|
|
|

|
| Sample: |
N-Dodecyl-β-D-Maltopyranoside 0, 102 kDa
Mce-family protein Mce4A monomer, 35 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
50mM Tris, 500mM NaCl, 10% Glycerol, 5mM DDM, 1mM Beta-ME, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Nov 28
|
Structural insights into the substrate-binding proteins Mce1A and Mce4A from Mycobacterium tuberculosis
IUCrJ 8(5) (2021)
Asthana P, Singh D, Pedersen J, Hynönen M, Sulu R, Murthy A, Laitaoja M, Jänis J, Riley L, Venkatesan R
|
| RgGuinier |
5.0 |
nm |
| Dmax |
19.1 |
nm |
| VolumePorod |
278 |
nm3 |
|
|
UniProt ID: Q15059 (25-416) Bromodomain-containing protein 3
|
|
|
|
| Sample: |
Bromodomain-containing protein 3 monomer, 44 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, and 2% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Sep 25
|
Multivalent nucleosome scaffolding by bromodomain and extraterminal domain tandem bromodomains.
J Biol Chem :108289 (2025)
Olp MD, Bursch KL, Wynia-Smith SL, Nuñez R, Goetz CJ, Jackson V, Smith BC
|
| RgGuinier |
4.7 |
nm |
| Dmax |
17.8 |
nm |
| VolumePorod |
85 |
nm3 |
|
|
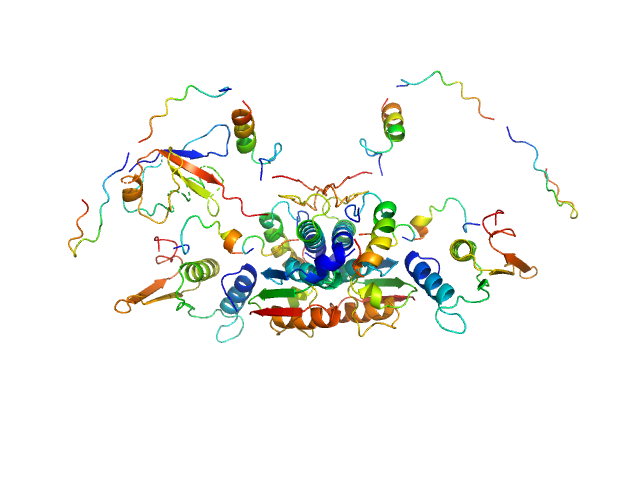
UniProt ID: Q9GZZ9 (57-346) Ubiquitin-like modifier-activating enzyme 5
UniProt ID: P61960 (1-83) Ubiquitin fold modifer 1
|
|
|
|
| Sample: |
Ubiquitin-like modifier-activating enzyme 5 dimer, 68 kDa Homo sapiens protein
Ubiquitin fold modifer 1 monomer, 9 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 50 mM NaCl, 2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Oct 29
|
Structure and dynamics of UBA5-UFM1 complex formation showing new insights in the UBA5 activation mechanism
Journal of Structural Biology :107796 (2021)
Fuchs S, Kikhney A, Schubert R, Kaiser C, Liebau E, Svergun D, Betzel C, Perbandt M
|
| RgGuinier |
3.1 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
106 |
nm3 |
|
|
UniProt ID: P02768 (None-None) Albumin
|
|
|
|
| Sample: |
Albumin monomer, 69 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM KCl, 2% glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2020 Dec 1
|
Albumin in patients with liver disease shows an altered conformation.
Commun Biol 4(1):731 (2021)
Paar M, Fengler VH, Rosenberg DJ, Krebs A, Stauber RE, Oettl K, Hammel M
|
| RgGuinier |
2.8 |
nm |
| Dmax |
8.3 |
nm |
|
|