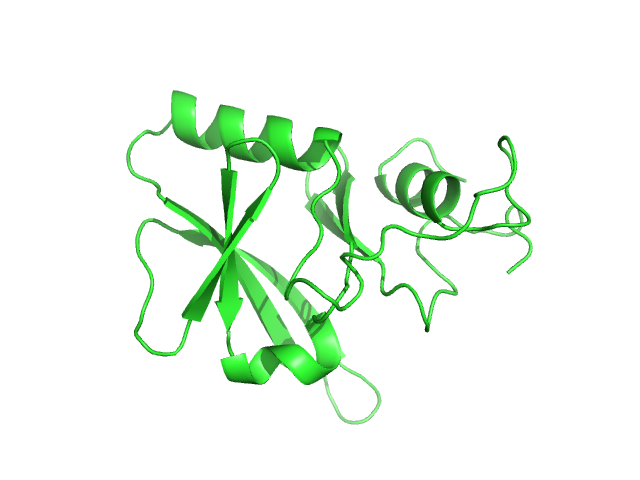
UniProt ID: P0DN75 (2-132) Iron-sulfur cluster assembly 1 homolog, mitochondrial
|
|
|
|
| Sample: |
Iron-sulfur cluster assembly 1 homolog, mitochondrial, 15 kDa Columba livia protein
|
| Buffer: |
20 mM Tris-HCl, 0.15 M NaCl, 10 mM 3-mercapto-1,2-propanediol, pH: 8 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2020 Feb 25
|
Magnetic field effects on the structure and molecular behavior of pigeon iron–sulfur protein
Protein Science 31(6) (2022)
Arai S, Shimizu R, Adachi M, Hirai M
|
|
|
UniProt ID: P9WJC1 (484-729) ESX-1 secretion-associated protein EspK
UniProt ID: P9WJD9 (7-278) ESX-1 secretion-associated protein EspB
|
|
|
|
| Sample: |
ESX-1 secretion-associated protein EspK monomer, 27 kDa Mycobacterium tuberculosis (strain … protein
ESX-1 secretion-associated protein EspB monomer, 30 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
20 mM Tris-HCl, 300 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Sep 12
|
The crystal structure of the EspB-EspK virulence factor-chaperone complex suggests an additional type VII secretion mechanism in M. tuberculosis.
J Biol Chem :102761 (2022)
Gijsbers A, Eymery M, Gao Y, Menart I, Vinciauskaite V, Siliqi D, Peters PJ, McCarthy A, Ravelli RBG
|
| RgGuinier |
4.1 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
113 |
nm3 |
|
|
UniProt ID: P74102 (2-317) Orange carotenoid-binding protein
|
|
|

|
| Sample: |
Orange carotenoid-binding protein monomer, 35 kDa Synechocystis sp. (strain … protein
|
| Buffer: |
50 mM Tris, 150 mM NaCL, pH: 7.4 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Mar 23
|
Oligomerization processes limit photoactivation and recovery of the orange carotenoid protein.
Biophys J 121(15):2849-2872 (2022)
Andreeva EA, Niziński S, Wilson A, Levantino M, De Zitter E, Munro R, Muzzopappa F, Thureau A, Zala N, Burdzinski G, Sliwa M, Kirilovsky D, Schirò G, Colletier JP
|
| RgGuinier |
2.3 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
58 |
nm3 |
|
|
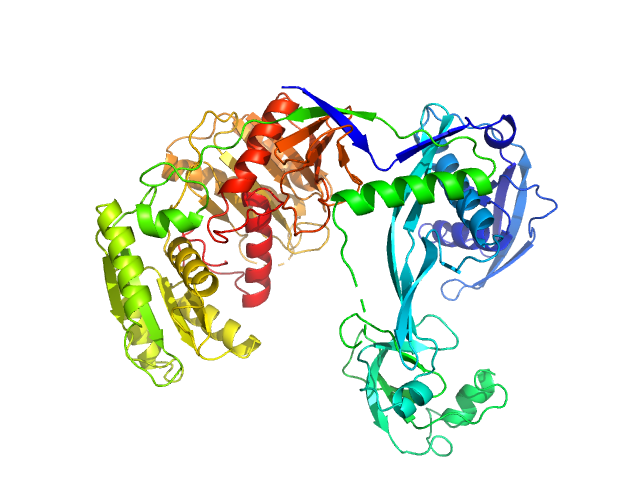
UniProt ID: Q746M7 (1-685) Piwi domain-containing protein
|
|
|
|
| Sample: |
Piwi domain-containing protein monomer, 77 kDa Thermus thermophilus (strain … protein
|
| Buffer: |
20 mM Tris–HCl, 250 mM NaCl, 2mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2021 May 7
|
Argonaute protein SAXS investigation
lirong zheng
|
| RgGuinier |
3.1 |
nm |
| Dmax |
9.7 |
nm |
| VolumePorod |
122 |
nm3 |
|
|
UniProt ID: P0DTC9 (44-180) Nucleoprotein
UniProt ID: None (None-None) Stem loop 4 with AU extension in the 5'-genomic end of SARS-CoV-2
|
|
|
|
| Sample: |
Nucleoprotein monomer, 15 kDa Severe acute respiratory … protein
Stem loop 4 with AU extension in the 5'-genomic end of SARS-CoV-2 monomer, 22 kDa Severe acute respiratory … RNA
|
| Buffer: |
25 mM potassium phosphate, 150 mM KCl, 2 mM TCEP, pH: 6.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Aug 16
|
The preference signature of the SARS-CoV-2 Nucleocapsid NTD for its 5'-genomic RNA elements.
Nat Commun 14(1):3331 (2023)
Korn SM, Dhamotharan K, Jeffries CM, Schlundt A
|
| RgGuinier |
3.4 |
nm |
| Dmax |
13.5 |
nm |
| VolumePorod |
51 |
nm3 |
|
|
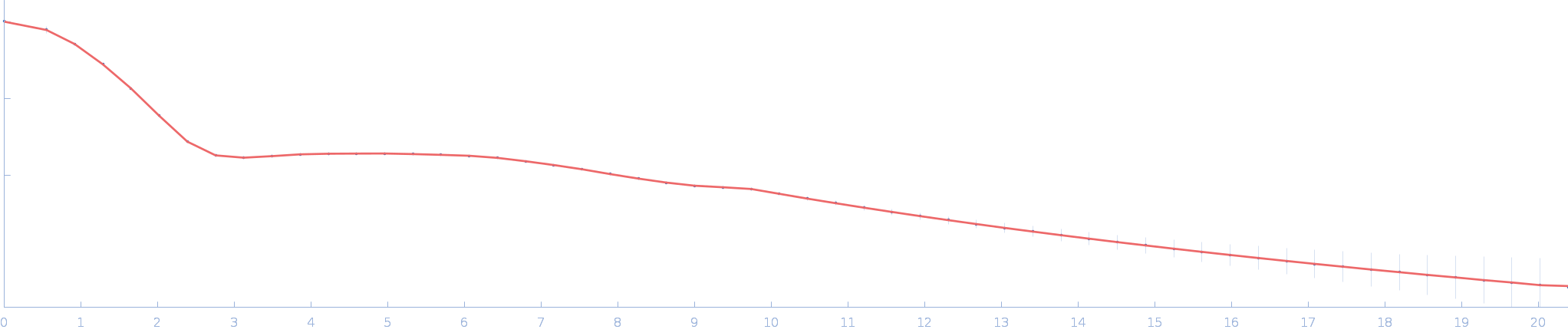
UniProt ID: P73129 (1-96) Ssr1698 protein (H21A)
|
|
|
|
| Sample: |
Ssr1698 protein (H21A) monomer, 11 kDa Synechocystis sp. (strain … protein
|
| Buffer: |
50 mM Hepes, 200 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at 16-ID (LiX), National Synchrotron Light Source II (NSLS-II) on 2022 Jun 10
|
A hemoprotein with a zinc-mirror heme site ties heme availability to carbon metabolism in cyanobacteria.
Nat Commun 15(1):3167 (2024)
Grosjean N, Yee EF, Kumaran D, Chopra K, Abernathy M, Biswas S, Byrnes J, Kreitler DF, Cheng JF, Ghosh A, Almo SC, Iwai M, Niyogi KK, Pakrasi HB, Sarangi R, van Dam H, Yang L, Blaby IK, Blaby-Haas CE
|
| RgGuinier |
1.6 |
nm |
| Dmax |
5.7 |
nm |
| VolumePorod |
12 |
nm3 |
|
|
UniProt ID: P20936 (174-1047) Ras GTPase-activating protein 1
UniProt ID: Q99704 (291-322) Docking protein 1
|
|
|
|
| Sample: |
Ras GTPase-activating protein 1 monomer, 101 kDa Homo sapiens protein
Docking protein 1 monomer, 3 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris pH 8, 150 mM NaCl, 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2021 Nov 4
|
Diverse p120RasGAP interactions with doubly phosphorylated partners EphB4, p190RhoGAP and Dok1
Journal of Biological Chemistry :105098 (2023)
Vish K, Stiegler A, Boggon T
|
| RgGuinier |
4.0 |
nm |
| Dmax |
14.6 |
nm |
| VolumePorod |
196 |
nm3 |
|
|
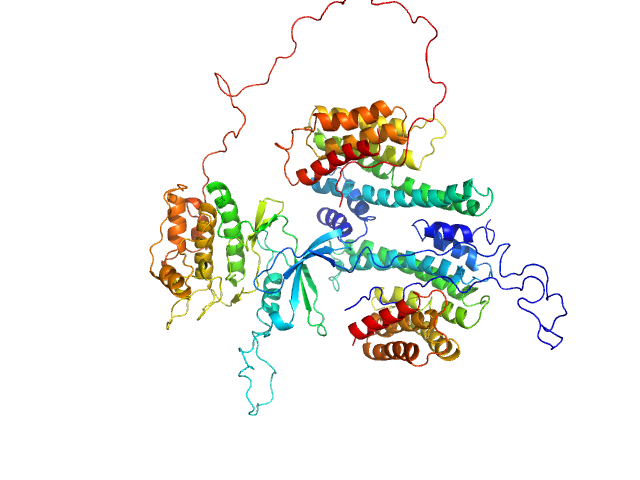
UniProt ID: P61981 (1-234) 14-3-3 protein gamma
UniProt ID: Q96RR4 (93-517) Calcium/calmodulin-dependent protein kinase kinase 2
|
|
|
|
| Sample: |
14-3-3 protein gamma dimer, 54 kDa Homo sapiens protein
Calcium/calmodulin-dependent protein kinase kinase 2 monomer, 48 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris-HCl, 150 mM NaCl, 1 mM TCEP, 3% (w/v) glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Oct 23
|
14-3-3 protein inhibits CaMKK1 by blocking the kinase active site with its last two C-terminal helices.
Protein Sci :e4805 (2023)
Petrvalska O, Honzejkova K, Koupilova N, Herman P, Obsilova V, Obsil T
|
| RgGuinier |
3.7 |
nm |
| Dmax |
13.2 |
nm |
| VolumePorod |
157 |
nm3 |
|
|
UniProt ID: P0DTC9 (1-245) Nucleoprotein
|
|
|
|
| Sample: |
Nucleoprotein monomer, 27 kDa Severe acute respiratory … protein
|
| Buffer: |
100 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 May 13
|
SARS-CoV-2 N-protein variants: N1-246 and IDL176-246
Guillem Hernandez
|
| RgGuinier |
3.3 |
nm |
| Dmax |
15.5 |
nm |
| VolumePorod |
56 |
nm3 |
|
|
UniProt ID: Q9WTS6-2 (343-2699) Isoform A0B0 of Teneurin-3
|
|
|
|
| Sample: |
Isoform A0B0 of Teneurin-3 dimer, 526 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 5 mM EDTA, pH: 7.8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2022 Sep 10
|
Alternative splicing controls teneurin-3 compact dimer formation for neuronal recognition
Nature Communications 15(1) (2024)
Gogou C, Beugelink J, Frias C, Kresik L, Jaroszynska N, Drescher U, Janssen B, Hindges R, Meijer D
|
| RgGuinier |
8.6 |
nm |
| Dmax |
33.0 |
nm |
| VolumePorod |
990 |
nm3 |
|
|