UniProt ID: P0DN75 (2-132) Iron-sulfur cluster assembly 1 homolog, mitochondrial
|
|
|
|
| Sample: |
Iron-sulfur cluster assembly 1 homolog, mitochondrial, 15 kDa Columba livia protein
|
| Buffer: |
20 mM Tris-HCl, 0.15 M NaCl, 10 mM 3-mercapto-1,2-propanediol, pH: 8 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2020 Nov 30
|
Magnetic field effects on the structure and molecular behavior of pigeon iron–sulfur protein
Protein Science 31(6) (2022)
Arai S, Shimizu R, Adachi M, Hirai M
|
| RgGuinier |
5.7 |
nm |
| Dmax |
18.9 |
nm |
|
|
UniProt ID: P74102 (2-317) Orange carotenoid-binding protein
|
|
|
|
| Sample: |
Orange carotenoid-binding protein, 105 kDa Synechocystis sp. (strain … protein
|
| Buffer: |
50 mM Tris, 150 mM NaCL, pH: 7.4 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Mar 23
|
Oligomerization processes limit photoactivation and recovery of the orange carotenoid protein.
Biophys J 121(15):2849-2872 (2022)
Andreeva EA, Niziński S, Wilson A, Levantino M, De Zitter E, Munro R, Muzzopappa F, Thureau A, Zala N, Burdzinski G, Sliwa M, Kirilovsky D, Schirò G, Colletier JP
|
| RgGuinier |
3.4 |
nm |
| Dmax |
19.0 |
nm |
| VolumePorod |
70 |
nm3 |
|
|
UniProt ID: P0DTC9 (1-245) Nucleoprotein (L223P, L227P, L230P)
|
|
|
|
| Sample: |
Nucleoprotein (L223P, L227P, L230P) monomer, 27 kDa Severe acute respiratory … protein
|
| Buffer: |
100 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 Jan 29
|
SARS-CoV-2 N-protein variants: N1-246 and IDL176-246
Guillem Hernandez
|
| RgGuinier |
4.4 |
nm |
| Dmax |
19.0 |
nm |
| VolumePorod |
68 |
nm3 |
|
|
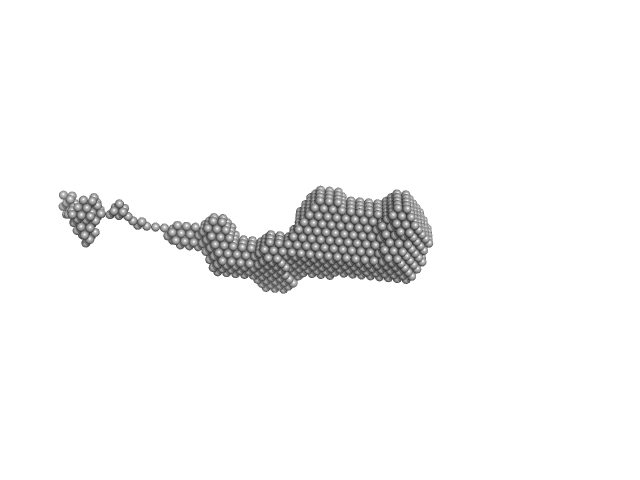
UniProt ID: Q9WTS6-3 (343-2706) Isoform A0B1 of Teneurin-3
|
|
|
|
| Sample: |
Isoform A0B1 of Teneurin-3 dimer, 545 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 2 mM CaCl2, pH: 7.8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2022 Sep 10
|
Alternative splicing controls teneurin-3 compact dimer formation for neuronal recognition
Nature Communications 15(1) (2024)
Gogou C, Beugelink J, Frias C, Kresik L, Jaroszynska N, Drescher U, Janssen B, Hindges R, Meijer D
|
| RgGuinier |
8.0 |
nm |
| Dmax |
32.0 |
nm |
| VolumePorod |
1053 |
nm3 |
|
|
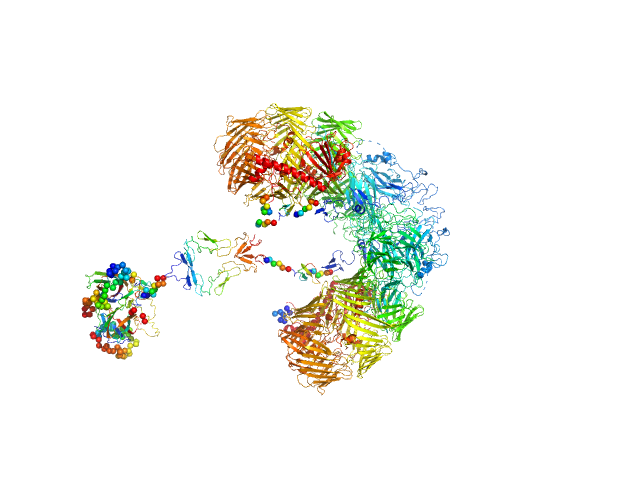
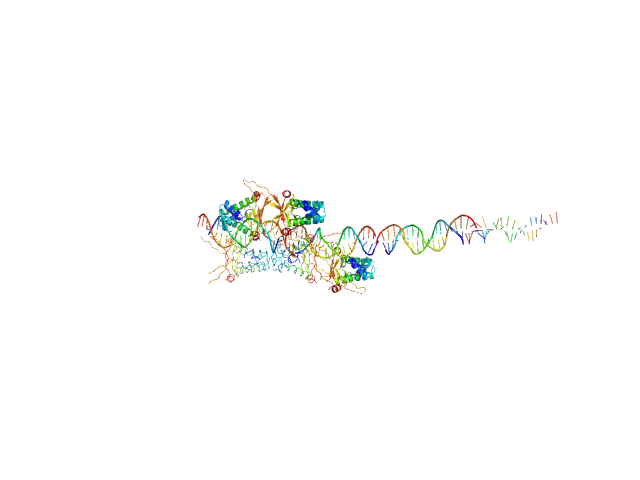
UniProt ID: None (None-None) 80bp_DNA Forward
UniProt ID: None (None-None) 80bp_DNA Reverse
UniProt ID: P0ACF0 (1-90) DNA-binding protein HU-alpha
|
|
|
|
| Sample: |
80bp_DNA Forward monomer, 25 kDa Escherichia coli DNA
80bp_DNA Reverse monomer, 25 kDa Escherichia coli DNA
DNA-binding protein HU-alpha decamer, 95 kDa Escherichia coli protein
|
| Buffer: |
10 mM Bis-Tris, 300 mM NaCl, pH: 5.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 May 27
|
Nucleoid remodeling during environmental adaptation is regulated by HU-dependent DNA bundling.
Nat Commun 11(1):2905 (2020)
Remesh SG, Verma SC, Chen JH, Ekman AA, Larabell CA, Adhya S, Hammel M
|
| RgGuinier |
6.4 |
nm |
| Dmax |
24.4 |
nm |
| VolumePorod |
268 |
nm3 |
|
|
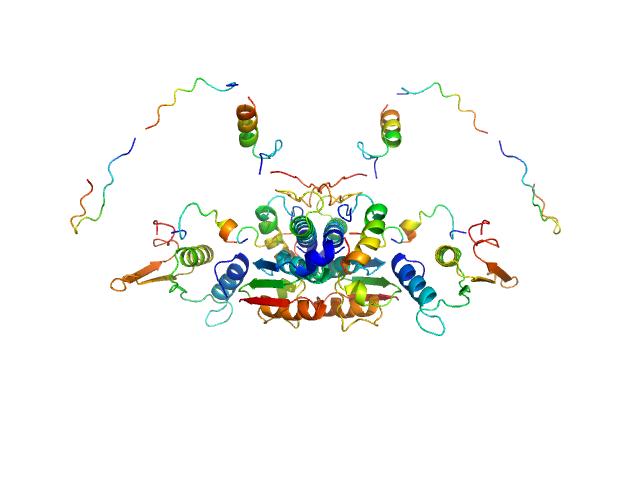
UniProt ID: Q9GZZ9 (57-346) Ubiquitin-like modifier-activating enzyme 5
|
|
|
|
| Sample: |
Ubiquitin-like modifier-activating enzyme 5 dimer, 68 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Jun 13
|
Structure and dynamics of UBA5-UFM1 complex formation showing new insights in the UBA5 activation mechanism
Journal of Structural Biology :107796 (2021)
Fuchs S, Kikhney A, Schubert R, Kaiser C, Liebau E, Svergun D, Betzel C, Perbandt M
|
| RgGuinier |
3.2 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
108 |
nm3 |
|
|
UniProt ID: P21675 (1373-1635) Transcription initiation factor TFIID subunit 1
|
|
|
|
| Sample: |
Transcription initiation factor TFIID subunit 1 monomer, 31 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, and 2% glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Sep 25
|
Multivalent nucleosome scaffolding by bromodomain and extraterminal domain tandem bromodomains.
J Biol Chem :108289 (2025)
Olp MD, Bursch KL, Wynia-Smith SL, Nuñez R, Goetz CJ, Jackson V, Smith BC
|
| RgGuinier |
2.9 |
nm |
| Dmax |
10.6 |
nm |
| VolumePorod |
65 |
nm3 |
|
|
UniProt ID: P02768 (None-None) Albumin
|
|
|
|
| Sample: |
Albumin monomer, 69 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM KCl, 2% glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2020 Dec 1
|
Albumin in patients with liver disease shows an altered conformation.
Commun Biol 4(1):731 (2021)
Paar M, Fengler VH, Rosenberg DJ, Krebs A, Stauber RE, Oettl K, Hammel M
|
| RgGuinier |
2.9 |
nm |
| Dmax |
9.0 |
nm |
|
|
UniProt ID: P0DN75 (2-132) Iron-sulfur cluster assembly 1 homolog, mitochondrial
|
|
|
|
| Sample: |
Iron-sulfur cluster assembly 1 homolog, mitochondrial, 15 kDa Columba livia protein
|
| Buffer: |
20 mM Tris-HCl, 0.15 M NaCl, 10 mM 3-mercapto-1,2-propanediol, pH: 8 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2020 Nov 30
|
Magnetic field effects on the structure and molecular behavior of pigeon iron–sulfur protein
Protein Science 31(6) (2022)
Arai S, Shimizu R, Adachi M, Hirai M
|
| RgGuinier |
5.7 |
nm |
| Dmax |
18.9 |
nm |
|
|
UniProt ID: P74102 (2-317) Orange carotenoid-binding protein
|
|
|

|
| Sample: |
Orange carotenoid-binding protein, 105 kDa Synechocystis sp. (strain … protein
|
| Buffer: |
50 mM Tris, 150 mM NaCL, pH: 7.4 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Mar 23
|
Oligomerization processes limit photoactivation and recovery of the orange carotenoid protein.
Biophys J 121(15):2849-2872 (2022)
Andreeva EA, Niziński S, Wilson A, Levantino M, De Zitter E, Munro R, Muzzopappa F, Thureau A, Zala N, Burdzinski G, Sliwa M, Kirilovsky D, Schirò G, Colletier JP
|
| RgGuinier |
4.8 |
nm |
| Dmax |
26.0 |
nm |
| VolumePorod |
120 |
nm3 |
|
|