UniProt ID: Q00623 (67-264) Apolipoprotein A-I
UniProt ID: None (None-None) 1,2-dimyristoyl-sn-glycero-3-phosphocholine
|
|
|
|
| Sample: |
Apolipoprotein A-I dimer, 50 kDa Mus musculus protein
1,2-dimyristoyl-sn-glycero-3-phosphocholine, 92 kDa
|
| Buffer: |
PBS in D2O, pH: 7.4 |
| Experiment: |
SAXS
data collected at SAXSLab Bio-Ganesha, Institute for Bioscience and Biotechnology Research, National Institute of Standards and Technology (IBBR/NIST) on 2015 Nov 24
|
Small-angle X-ray and neutron scattering demonstrates that cell-free expression produces properly formed disc-shaped nanolipoprotein particles.
Protein Sci 27(3):780-789 (2018)
Cleveland TE 4th, He W, Evans AC, Fischer NO, Lau EY, Coleman MA, Butler P
|
| RgGuinier |
4.8 |
nm |
| Dmax |
12.0 |
nm |
|
|
UniProt ID: Q00623 (67-264) Apolipoprotein A-I
UniProt ID: None (None-None) 1,2-dimyristoyl-d54-sn-glycero-3-phosphocholine
|
|
|
|
| Sample: |
Apolipoprotein A-I dimer, 50 kDa Mus musculus protein
1,2-dimyristoyl-d54-sn-glycero-3-phosphocholine, 99 kDa
|
| Buffer: |
PBS in D2O, pH: 7.4 |
| Experiment: |
SANS
data collected at NG7, NIST Center for High Resolution Neutron Scattering (CHRNS) on 2016 Apr 11
|
Small-angle X-ray and neutron scattering demonstrates that cell-free expression produces properly formed disc-shaped nanolipoprotein particles.
Protein Sci 27(3):780-789 (2018)
Cleveland TE 4th, He W, Evans AC, Fischer NO, Lau EY, Coleman MA, Butler P
|
| RgGuinier |
4.3 |
nm |
| Dmax |
10.7 |
nm |
|
|
UniProt ID: Q00623 (67-264) Apolipoprotein A-I
UniProt ID: None (None-None) 1,2-dimyristoyl-sn-glycero-3-phosphocholine
|
|
|
|
| Sample: |
Apolipoprotein A-I dimer, 50 kDa Mus musculus protein
1,2-dimyristoyl-sn-glycero-3-phosphocholine, 92 kDa
|
| Buffer: |
PBS in 42% D2O, pH: 7.4 |
| Experiment: |
SANS
data collected at NGB 30m SANS, NIST Center for High Resolution Neutron Scattering (CHRNS) on 2015 Dec 11
|
Small-angle X-ray and neutron scattering demonstrates that cell-free expression produces properly formed disc-shaped nanolipoprotein particles.
Protein Sci 27(3):780-789 (2018)
Cleveland TE 4th, He W, Evans AC, Fischer NO, Lau EY, Coleman MA, Butler P
|
| RgGuinier |
2.9 |
nm |
| Dmax |
7.3 |
nm |
|
|
UniProt ID: None (None-None) 1,2-dimyristoyl-sn-glycero-3-phosphocholine
UniProt ID: Q00623 (67-264) Apolipoprotein A-I
|
|
|
|
| Sample: |
1,2-dimyristoyl-sn-glycero-3-phosphocholine, 92 kDa
Apolipoprotein A-I dimer, 52 kDa Mus musculus protein
|
| Buffer: |
PBS in D2O, pH: 7.4 |
| Experiment: |
SAXS
data collected at SAXSLab Bio-Ganesha, Institute for Bioscience and Biotechnology Research, National Institute of Standards and Technology (IBBR/NIST) on 2015 Nov 24
|
Small-angle X-ray and neutron scattering demonstrates that cell-free expression produces properly formed disc-shaped nanolipoprotein particles.
Protein Sci 27(3):780-789 (2018)
Cleveland TE 4th, He W, Evans AC, Fischer NO, Lau EY, Coleman MA, Butler P
|
| RgGuinier |
4.7 |
nm |
| Dmax |
12.7 |
nm |
|
|
UniProt ID: None (None-None) 1,2-dimyristoyl-sn-glycero-3-phosphocholine
UniProt ID: Q00623 (67-264) Apolipoprotein A-I
|
|
|
|
| Sample: |
1,2-dimyristoyl-sn-glycero-3-phosphocholine, 92 kDa
Apolipoprotein A-I dimer, 52 kDa Mus musculus protein
|
| Buffer: |
PBS in D2O, pH: 7.4 |
| Experiment: |
SAXS
data collected at SAXSLab Bio-Ganesha, Institute for Bioscience and Biotechnology Research, National Institute of Standards and Technology (IBBR/NIST) on 2015 Nov 24
|
Small-angle X-ray and neutron scattering demonstrates that cell-free expression produces properly formed disc-shaped nanolipoprotein particles.
Protein Sci 27(3):780-789 (2018)
Cleveland TE 4th, He W, Evans AC, Fischer NO, Lau EY, Coleman MA, Butler P
|
| RgGuinier |
4.4 |
nm |
| Dmax |
12.7 |
nm |
|
|
UniProt ID: None (None-None) 1,2-dimyristoyl-sn-glycero-3-phosphocholine
UniProt ID: Q00623 (67-264) Apolipoprotein A-I
|
|
|
|
| Sample: |
1,2-dimyristoyl-sn-glycero-3-phosphocholine, 92 kDa
Apolipoprotein A-I dimer, 52 kDa Mus musculus protein
|
| Buffer: |
PBS in D2O, pH: 7.4 |
| Experiment: |
SANS
data collected at NG7, NIST Center for High Resolution Neutron Scattering (CHRNS) on 2015 Nov 25
|
Small-angle X-ray and neutron scattering demonstrates that cell-free expression produces properly formed disc-shaped nanolipoprotein particles.
Protein Sci 27(3):780-789 (2018)
Cleveland TE 4th, He W, Evans AC, Fischer NO, Lau EY, Coleman MA, Butler P
|
| RgGuinier |
3.5 |
nm |
| Dmax |
12.5 |
nm |
|
|
UniProt ID: None (None-None) 1,2-dimyristoyl-sn-glycero-3-phosphocholine
UniProt ID: Q00623 (67-264) Apolipoprotein A-I
|
|
|
|
| Sample: |
1,2-dimyristoyl-sn-glycero-3-phosphocholine, 92 kDa
Apolipoprotein A-I dimer, 52 kDa Mus musculus protein
|
| Buffer: |
PBS in D2O, pH: 7.4 |
| Experiment: |
SANS
data collected at NG7, NIST Center for High Resolution Neutron Scattering (CHRNS) on 2015 Nov 25
|
Small-angle X-ray and neutron scattering demonstrates that cell-free expression produces properly formed disc-shaped nanolipoprotein particles.
Protein Sci 27(3):780-789 (2018)
Cleveland TE 4th, He W, Evans AC, Fischer NO, Lau EY, Coleman MA, Butler P
|
| RgGuinier |
3.3 |
nm |
| Dmax |
10.0 |
nm |
|
|
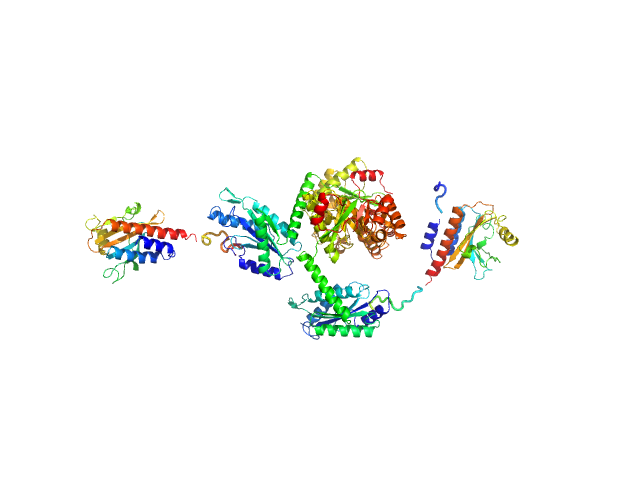
UniProt ID: A0QUG3 (None-None) Sensory box/response regulator
|
|
|
|
| Sample: |
Sensory box/response regulator dimer, 136 kDa Mycobacterium smegmatis (strain … protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, 5% glycerol, 2 mM MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2016 Jun 26
|
The GDP-switched GAF domain of DcpA modulates the concerted synthesis/hydrolysis of c-di-GMP in Mycobacterium smegmatis.
Biochem J 475(7):1295-1308 (2018)
Chen HJ, Li N, Luo Y, Jiang YL, Zhou CZ, Chen Y, Li Q
|
| RgGuinier |
5.0 |
nm |
| Dmax |
20.0 |
nm |
| VolumePorod |
299 |
nm3 |
|
|
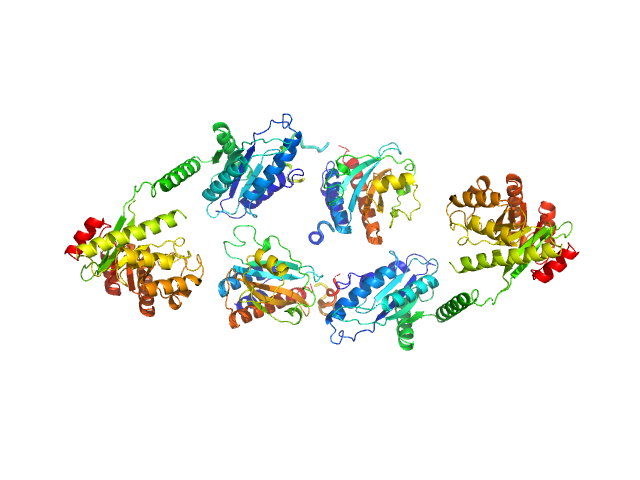
UniProt ID: A0QUG3 (None-None) Sensory box/response regulator
|
|
|
|
| Sample: |
Sensory box/response regulator dimer, 136 kDa Mycobacterium smegmatis (strain … protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, 5% glycerol, 2 mM MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2016 Jun 26
|
The GDP-switched GAF domain of DcpA modulates the concerted synthesis/hydrolysis of c-di-GMP in Mycobacterium smegmatis.
Biochem J 475(7):1295-1308 (2018)
Chen HJ, Li N, Luo Y, Jiang YL, Zhou CZ, Chen Y, Li Q
|
| RgGuinier |
4.8 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
271 |
nm3 |
|
|
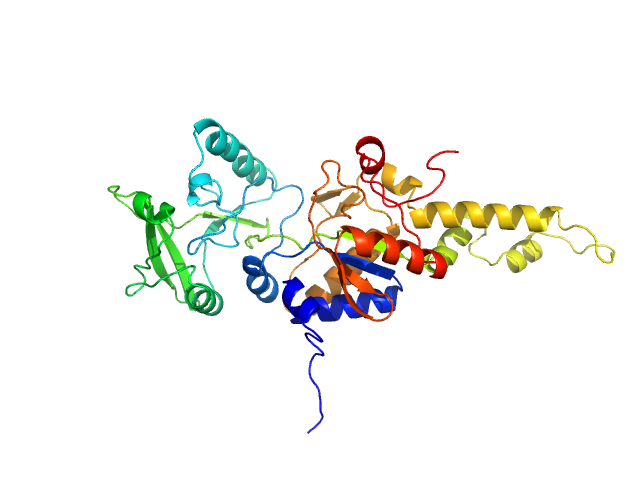
UniProt ID: P0DKX7 (1-364) Bifunctional hemolysin/adenylate cyclase
|
|
|
|
| Sample: |
Bifunctional hemolysin/adenylate cyclase monomer, 39 kDa Bordetella pertussis protein
|
| Buffer: |
20 mM Hepes, 150 mM NaCl, 4 mM CaCl2, pH: 7.4 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2015 Jun 19
|
Calmodulin fishing with a structurally disordered bait triggers CyaA catalysis.
PLoS Biol 15(12):e2004486 (2017)
O'Brien DP, Durand D, Voegele A, Hourdel V, Davi M, Chamot-Rooke J, Vachette P, Brier S, Ladant D, Chenal A
|
| RgGuinier |
2.6 |
nm |
| Dmax |
9.1 |
nm |
| VolumePorod |
60 |
nm3 |
|
|