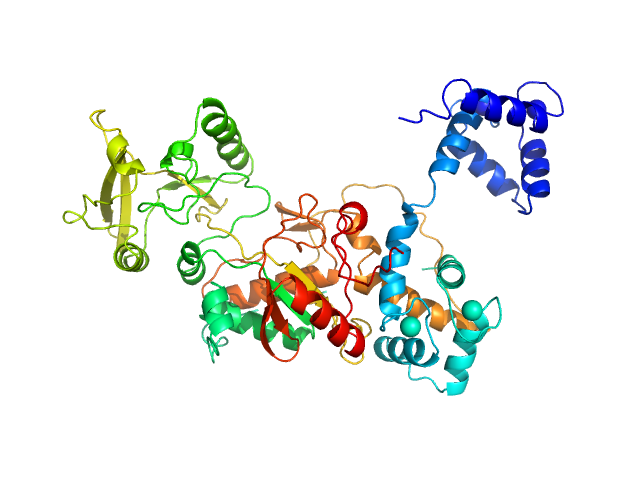
UniProt ID: P0DKX7 (1-364) Bifunctional hemolysin/adenylate cyclase
UniProt ID: P62158 (None-None) Calmodulin
|
|
|
|
| Sample: |
Bifunctional hemolysin/adenylate cyclase monomer, 39 kDa Bordetella pertussis protein
Calmodulin monomer, 17 kDa Homo sapiens protein
|
| Buffer: |
20 mM Hepes, 150 mM NaCl, 4 mM CaCl2, pH: 7.4 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2015 Jun 19
|
Calmodulin fishing with a structurally disordered bait triggers CyaA catalysis.
PLoS Biol 15(12):e2004486 (2017)
O'Brien DP, Durand D, Voegele A, Hourdel V, Davi M, Chamot-Rooke J, Vachette P, Brier S, Ladant D, Chenal A
|
| RgGuinier |
2.9 |
nm |
| Dmax |
9.8 |
nm |
| VolumePorod |
78 |
nm3 |
|
|
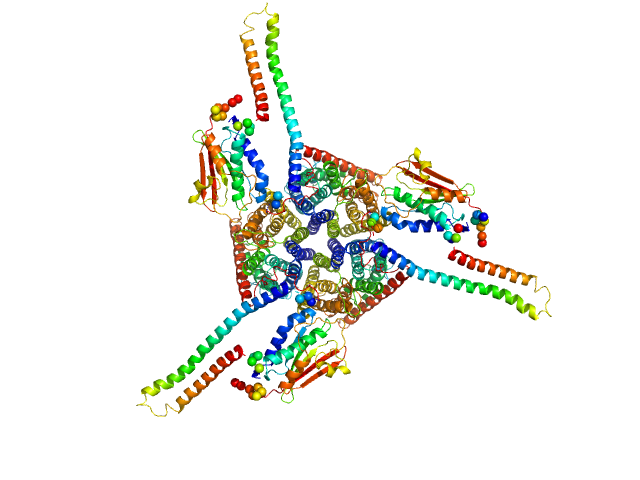
UniProt ID: Q1Q357 (None-None) ammonium sensor/transducer
|
|
|
|
| Sample: |
Ammonium sensor/transducer trimer, 227 kDa Candidatus Kuenenia stuttgartiensis protein
|
| Buffer: |
20 mM Tris/HCl,100 mM NaCl, 5%(w/v) glycerol, 0.09%(w/v) CYMAL-5, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Mar 1
|
Signaling ammonium across membranes through an ammonium sensor histidine kinase.
Nat Commun 9(1):164 (2018)
Pflüger T, Hernández CF, Lewe P, Frank F, Mertens H, Svergun D, Baumstark MW, Lunin VY, Jetten MSM, Andrade SLA
|
| RgGuinier |
4.8 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
511 |
nm3 |
|
|
UniProt ID: Q1Q357 (None-None) Ammonium transporter histidine kinase domain
|
|
|
|
| Sample: |
Ammonium transporter histidine kinase domain monomer, 30 kDa Candidatus Kuenenia stuttgartiensis protein
|
| Buffer: |
20 mM Tris/HCl,100 mM NaCl, 5%(w/v) glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Mar 2
|
Signaling ammonium across membranes through an ammonium sensor histidine kinase.
Nat Commun 9(1):164 (2018)
Pflüger T, Hernández CF, Lewe P, Frank F, Mertens H, Svergun D, Baumstark MW, Lunin VY, Jetten MSM, Andrade SLA
|
| RgGuinier |
2.7 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
71 |
nm3 |
|
|
UniProt ID: Q1Q357 (None-None) Ammonium transporter histidine kinase domain
|
|
|
|
| Sample: |
Ammonium transporter histidine kinase domain monomer, 30 kDa Candidatus Kuenenia stuttgartiensis protein
|
| Buffer: |
20 mM Tris/HCl,100 mM NaCl, 5%(w/v) glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Mar 2
|
Signaling ammonium across membranes through an ammonium sensor histidine kinase.
Nat Commun 9(1):164 (2018)
Pflüger T, Hernández CF, Lewe P, Frank F, Mertens H, Svergun D, Baumstark MW, Lunin VY, Jetten MSM, Andrade SLA
|
| RgGuinier |
2.3 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
53 |
nm3 |
|
|
UniProt ID: Q1Q357 (None-None) Ammonium transporter histidine kinase domain
|
|
|
|
| Sample: |
Ammonium transporter histidine kinase domain monomer, 30 kDa Candidatus Kuenenia stuttgartiensis protein
|
| Buffer: |
20 mM Tris/HCl,100 mM NaCl, 5%(w/v) glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Mar 2
|
Signaling ammonium across membranes through an ammonium sensor histidine kinase.
Nat Commun 9(1):164 (2018)
Pflüger T, Hernández CF, Lewe P, Frank F, Mertens H, Svergun D, Baumstark MW, Lunin VY, Jetten MSM, Andrade SLA
|
| RgGuinier |
2.2 |
nm |
| Dmax |
7.7 |
nm |
| VolumePorod |
53 |
nm3 |
|
|
UniProt ID: None (None-None) Designed Ankyrin Repeat Protein D1
UniProt ID: O00139 (153-553) Kinesin-like protein KIF2A
UniProt ID: P81947 (None-None) Tubulin alpha-1B chain
UniProt ID: Q6B856 (None-None) Tubulin beta-2B chain
|
|
|

|
| Sample: |
Designed Ankyrin Repeat Protein D1 monomer, 18 kDa synthetic construct protein
Kinesin-like protein KIF2A monomer, 48 kDa Homo sapiens protein
Tubulin alpha-1B chain dimer, 100 kDa Bos taurus protein
Tubulin beta-2B chain dimer, 100 kDa Bos taurus protein
|
| Buffer: |
HEPES 20 mM, MgCl2 1mM, NaCl 150mM, pH: 7.2 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2016 May 16
|
Ternary complex of Kif2A-bound tandem tubulin heterodimers represents a kinesin-13-mediated microtubule depolymerization reaction intermediate.
Nat Commun 9(1):2628 (2018)
Trofimova D, Paydar M, Zara A, Talje L, Kwok BH, Allingham JS
|
| RgGuinier |
5.4 |
nm |
| Dmax |
19.5 |
nm |
| VolumePorod |
374 |
nm3 |
|
|
UniProt ID: Q1Q357 (None-None) ammonium sensor/transducer
|
|
|
|
| Sample: |
Ammonium sensor/transducer trimer, 227 kDa Candidatus Kuenenia stuttgartiensis protein
|
| Buffer: |
20 mM Tris/HCl,100 mM NaCl, 5%(w/v) glycerol, 0.09%(w/v) CYMAL-5, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Mar 2
|
Signaling ammonium across membranes through an ammonium sensor histidine kinase.
Nat Commun 9(1):164 (2018)
Pflüger T, Hernández CF, Lewe P, Frank F, Mertens H, Svergun D, Baumstark MW, Lunin VY, Jetten MSM, Andrade SLA
|
| RgGuinier |
4.9 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
482 |
nm3 |
|
|
UniProt ID: A6Q0K5 (28-107) Calvin cycle protein CP12, chloroplastic
|
|
|
|
| Sample: |
Calvin cycle protein CP12, chloroplastic monomer, 11 kDa Chlamydomonas reinhardtii protein
|
| Buffer: |
50 mM phosphate buffer, 50 mM NaCl, 20 mM oxidized DTT, pH: 6.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2015 Nov 3
|
Cryptic Disorder Out of Disorder: Encounter between Conditionally Disordered CP12 and Glyceraldehyde-3-Phosphate Dehydrogenase.
J Mol Biol 430(8):1218-1234 (2018)
Launay H, Barré P, Puppo C, Zhang Y, Maneville S, Gontero B, Receveur-Bréchot V
|
| RgGuinier |
2.3 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
22 |
nm3 |
|
|
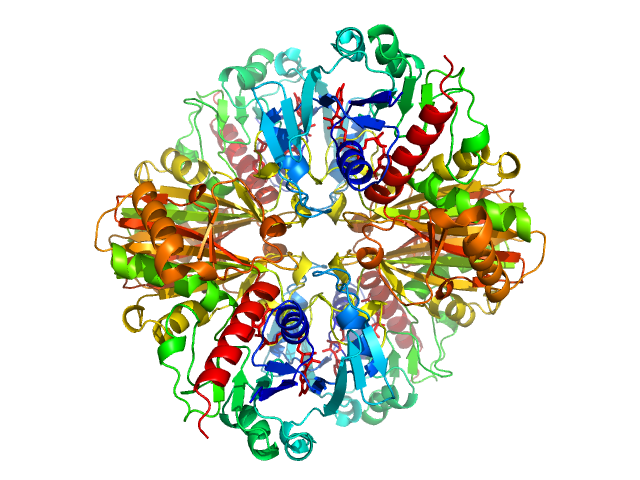
UniProt ID: A8HP84 (None-None) Glyceraldehyde-3-phosphate dehydrogenase
|
|
|
|
| Sample: |
Glyceraldehyde-3-phosphate dehydrogenase tetramer, 148 kDa Chlamydomonas reinhardtii protein
|
| Buffer: |
30 mM Tris, 4 mM EDTA, 100 µM NAD, 5 mM free cysteine, pH: 7.9 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2011 Feb 7
|
Cryptic Disorder Out of Disorder: Encounter between Conditionally Disordered CP12 and Glyceraldehyde-3-Phosphate Dehydrogenase.
J Mol Biol 430(8):1218-1234 (2018)
Launay H, Barré P, Puppo C, Zhang Y, Maneville S, Gontero B, Receveur-Bréchot V
|
| RgGuinier |
3.2 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
206 |
nm3 |
|
|
UniProt ID: A6Q0K5 (28-107) Calvin cycle protein CP12, chloroplastic
UniProt ID: A8HP84 (None-None) Glyceraldehyde-3-phosphate dehydrogenase
|
|
|
|
| Sample: |
Calvin cycle protein CP12, chloroplastic monomer, 11 kDa Chlamydomonas reinhardtii protein
Glyceraldehyde-3-phosphate dehydrogenase tetramer, 148 kDa Chlamydomonas reinhardtii protein
|
| Buffer: |
30 mM Tris, 4 mM EDTA, 100 µM NAD, 5 mM free cysteine, pH: 7.9 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2010 Dec 8
|
Cryptic Disorder Out of Disorder: Encounter between Conditionally Disordered CP12 and Glyceraldehyde-3-Phosphate Dehydrogenase.
J Mol Biol 430(8):1218-1234 (2018)
Launay H, Barré P, Puppo C, Zhang Y, Maneville S, Gontero B, Receveur-Bréchot V
|
| RgGuinier |
3.8 |
nm |
| Dmax |
19.0 |
nm |
| VolumePorod |
251 |
nm3 |
|
|