UniProt ID: P41365 (26-342) Lipase B from Pseudozyma antarctica
|
|
|
|
| Sample: |
Lipase B from Pseudozyma antarctica, 33 kDa Moesziomyces antarcticus protein
|
| Buffer: |
100 mM NaCl, 20 mM Na2HPO4, 10 mM DTT, pH: 6 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Jul 29
|
Machine Learning Methods for X-Ray Scattering Data Analysis from Biomacromolecular Solutions.
Biophys J 114(11):2485-2492 (2018)
Franke D, Jeffries CM, Svergun DI
|
|
|
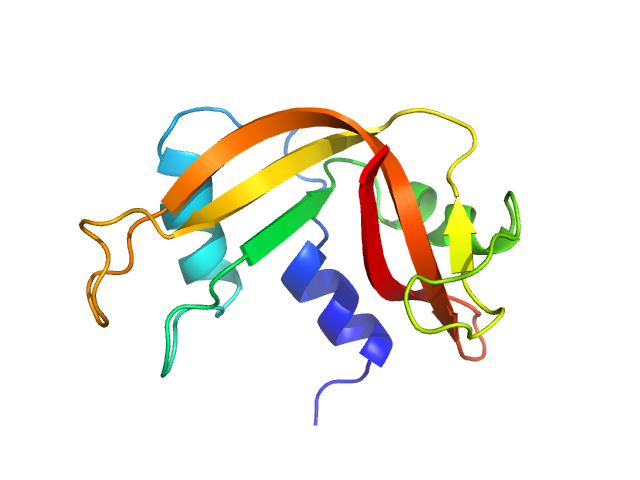
UniProt ID: P61823 (None-None) Ribonuclease pancreatic
|
|
|
|
| Sample: |
Ribonuclease pancreatic monomer, 16 kDa Bos taurus protein
|
| Buffer: |
phosphate buffered saline (PBS), pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Jul 29
|
Machine Learning Methods for X-Ray Scattering Data Analysis from Biomacromolecular Solutions.
Biophys J 114(11):2485-2492 (2018)
Franke D, Jeffries CM, Svergun DI
|
| RgGuinier |
1.6 |
nm |
| Dmax |
5.6 |
nm |
| VolumePorod |
16 |
nm3 |
|
|
UniProt ID: P61823 (None-None) Ribonuclease pancreatic
|
|
|
|
| Sample: |
Ribonuclease pancreatic monomer, 16 kDa Bos taurus protein
|
| Buffer: |
10 mM HCl, pH: 1 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Jul 29
|
Machine Learning Methods for X-Ray Scattering Data Analysis from Biomacromolecular Solutions.
Biophys J 114(11):2485-2492 (2018)
Franke D, Jeffries CM, Svergun DI
|
| RgGuinier |
2.3 |
nm |
| Dmax |
9.0 |
nm |
|
|
UniProt ID: P02769 (25-607) Bovine serum albumin
|
|
|
|
| Sample: |
Bovine serum albumin, 66 kDa Bos taurus protein
|
| Buffer: |
50 mM HEPES, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Sep 25
|
Machine Learning Methods for X-Ray Scattering Data Analysis from Biomacromolecular Solutions.
Biophys J 114(11):2485-2492 (2018)
Franke D, Jeffries CM, Svergun DI
|
| RgGuinier |
3.0 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
117 |
nm3 |
|
|
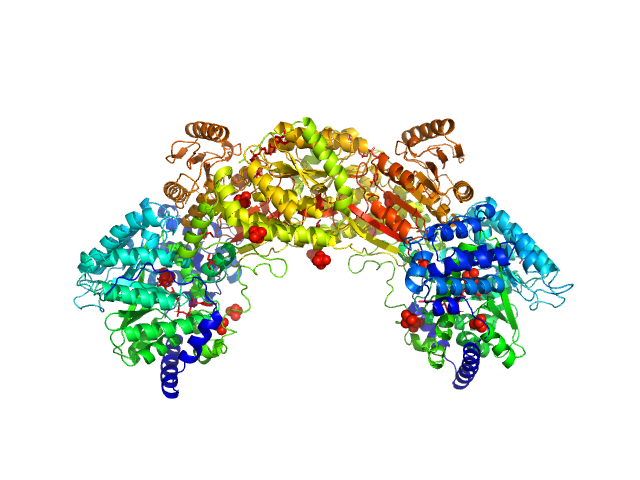
UniProt ID: Q89E26 (None-None) Bifunctional protein PutA
|
|
|
|
| Sample: |
Bifunctional protein PutA dimer, 215 kDa Bradyrhizobium diazoefficiens protein
|
| Buffer: |
50 mM Tris, 50 mM NaCl, 0.5 mM TCEP, 5% (v/v) glycerol, pH: 7.8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2017 Jul 16
|
Redox Modulation of Oligomeric State in Proline Utilization A.
Biophys J 114(12):2833-2843 (2018)
Korasick DA, Campbell AC, Christgen SL, Chakravarthy S, White TA, Becker DF, Tanner JJ
|
| RgGuinier |
4.6 |
nm |
| Dmax |
14.4 |
nm |
| VolumePorod |
324 |
nm3 |
|
|
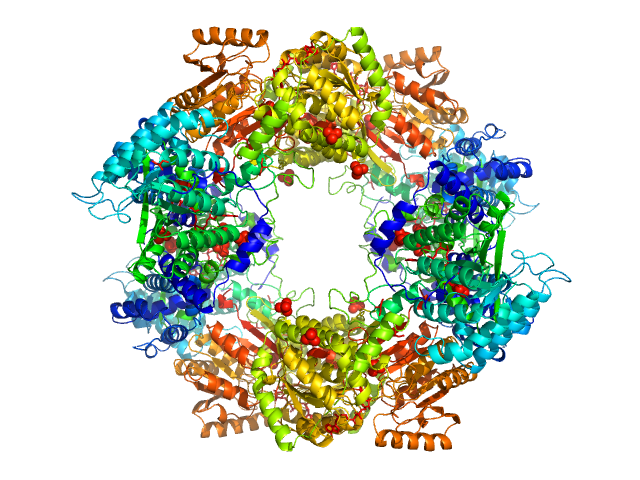
UniProt ID: Q89E26 (None-None) Bifunctional protein PutA
|
|
|
|
| Sample: |
Bifunctional protein PutA tetramer, 430 kDa Bradyrhizobium diazoefficiens protein
|
| Buffer: |
50 mM Tris, 50 mM NaCl, 0.5 mM TCEP, 5% (v/v) glycerol, pH: 7.8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2017 Jul 16
|
Redox Modulation of Oligomeric State in Proline Utilization A.
Biophys J 114(12):2833-2843 (2018)
Korasick DA, Campbell AC, Christgen SL, Chakravarthy S, White TA, Becker DF, Tanner JJ
|
| RgGuinier |
5.2 |
nm |
| Dmax |
14.2 |
nm |
| VolumePorod |
582 |
nm3 |
|
|
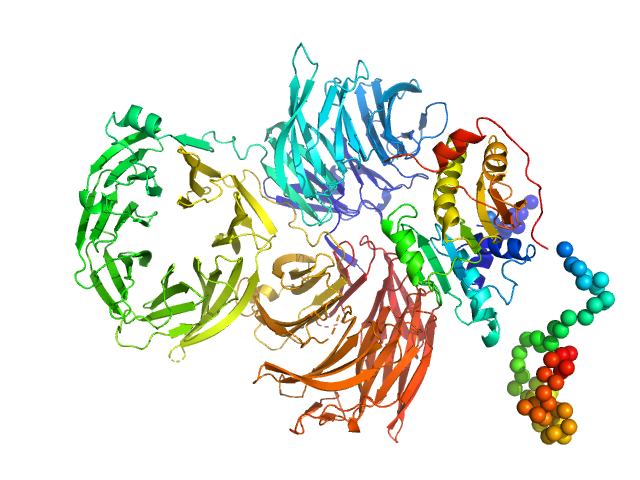
UniProt ID: P38238 (1-310) Trm7: tRNA (cytidine(32)/guanosine(34)-2'-O)-methyltransferase
UniProt ID: Q08924 (1-1013) Trm734: Regulator of Ty1 transposition protein 10
|
|
|
|
| Sample: |
Trm7: tRNA (cytidine(32)/guanosine(34)-2'-O)-methyltransferase monomer, 36 kDa Saccharomyces cerevisiae protein
Trm734: Regulator of Ty1 transposition protein 10 monomer, 116 kDa Saccharomyces cerevisiae protein
|
| Buffer: |
50 mM HEPES, 200 mM KCl, 5% v/v Glycerol, 10mM β-mercaptoethanol, pH: 8 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2015 Dec 16
|
Structure of tRNA methyltransferase complex of Trm7 and Trm734 reveals a novel binding interface for tRNA recognition.
Nucleic Acids Res (2019)
Hirata A, Okada K, Yoshii K, Shiraishi H, Saijo S, Yonezawa K, Shimizu N, Hori H
|
| RgGuinier |
3.8 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
218 |
nm3 |
|
|
UniProt ID: Q6Q271 (None-None) p-hydroxyphenylacetate 3-hydroxylase, reductase component
|
|
|
|
| Sample: |
P-hydroxyphenylacetate 3-hydroxylase, reductase component dimer, 71 kDa Acinetobacter baumannii protein
|
| Buffer: |
50 mM MOPS, 0.5 mM EDTA, 1 mM DTT, and 5% glycerol, pH: 7 |
| Experiment: |
SAXS
data collected at BL1.3W, Synchrotron Light Research Institute (SLRI) on 2017 Mar 7
|
Crystal structure of the flavin reductase of Acinetobacter baumannii p-hydroxyphenylacetate 3-hydroxylase (HPAH) and identification of amino acid residues underlying its regulation by aromatic ligands.
Arch Biochem Biophys 653:24-38 (2018)
Yuenyao A, Petchyam N, Kamonsutthipaijit N, Chaiyen P, Pakotiprapha D
|
| RgGuinier |
2.4 |
nm |
| Dmax |
6.9 |
nm |
| VolumePorod |
94 |
nm3 |
|
|
UniProt ID: Q6Q271 (None-None) p-hydroxyphenylacetate 3-hydroxylase, reductase component
|
|
|
|
| Sample: |
P-hydroxyphenylacetate 3-hydroxylase, reductase component dimer, 71 kDa Acinetobacter baumannii protein
|
| Buffer: |
50 mM MOPS, 0.5 mM EDTA, 1 mM DTT, and 5% glycerol, pH: 7 |
| Experiment: |
SAXS
data collected at BL1.3W, Synchrotron Light Research Institute (SLRI) on 2017 Mar 7
|
Crystal structure of the flavin reductase of Acinetobacter baumannii p-hydroxyphenylacetate 3-hydroxylase (HPAH) and identification of amino acid residues underlying its regulation by aromatic ligands.
Arch Biochem Biophys 653:24-38 (2018)
Yuenyao A, Petchyam N, Kamonsutthipaijit N, Chaiyen P, Pakotiprapha D
|
| RgGuinier |
2.4 |
nm |
| Dmax |
7.1 |
nm |
| VolumePorod |
95 |
nm3 |
|
|
UniProt ID: Q6Q271 (None-None) p-hydroxyphenylacetate 3-hydroxylase, reductase component
|
|
|
|
| Sample: |
P-hydroxyphenylacetate 3-hydroxylase, reductase component dimer, 71 kDa Acinetobacter baumannii protein
|
| Buffer: |
50 mM MOPS, 0.5 mM EDTA, 1 mM DTT, and 5% glycerol, pH: 7 |
| Experiment: |
SAXS
data collected at BL1.3W, Synchrotron Light Research Institute (SLRI) on 2017 Mar 7
|
Crystal structure of the flavin reductase of Acinetobacter baumannii p-hydroxyphenylacetate 3-hydroxylase (HPAH) and identification of amino acid residues underlying its regulation by aromatic ligands.
Arch Biochem Biophys 653:24-38 (2018)
Yuenyao A, Petchyam N, Kamonsutthipaijit N, Chaiyen P, Pakotiprapha D
|
| RgGuinier |
2.4 |
nm |
| Dmax |
7.2 |
nm |
| VolumePorod |
95 |
nm3 |
|
|