UniProt ID: Q6Q271 (None-None) p-hydroxyphenylacetate 3-hydroxylase (HPAH), reductase component E251A mutant
|
|
|
|
| Sample: |
P-hydroxyphenylacetate 3-hydroxylase (HPAH), reductase component E251A mutant dimer, 71 kDa Acinetobacter baumannii protein
|
| Buffer: |
50 mM MOPS, 0.5 mM EDTA, 1 mM DTT, 50 mM NaCl, 10 % glycerol, pH: 7 |
| Experiment: |
SAXS
data collected at BL1.3W, Synchrotron Light Research Institute (SLRI) on 2018 Apr 25
|
Crystal structure of the flavin reductase of Acinetobacter baumannii p-hydroxyphenylacetate 3-hydroxylase (HPAH) and identification of amino acid residues underlying its regulation by aromatic ligands.
Arch Biochem Biophys 653:24-38 (2018)
Yuenyao A, Petchyam N, Kamonsutthipaijit N, Chaiyen P, Pakotiprapha D
|
| RgGuinier |
2.7 |
nm |
| Dmax |
8.9 |
nm |
| VolumePorod |
93 |
nm3 |
|
|
UniProt ID: Q6Q271 (None-None) p-hydroxyphenylacetate 3-hydroxylase (HPAH), reductase component E251A mutant
|
|
|
|
| Sample: |
P-hydroxyphenylacetate 3-hydroxylase (HPAH), reductase component E251A mutant dimer, 71 kDa Acinetobacter baumannii protein
|
| Buffer: |
50 mM MOPS, 0.5 mM EDTA, 1 mM DTT, 50 mM NaCl, 10 % glycerol, pH: 7 |
| Experiment: |
SAXS
data collected at BL1.3W, Synchrotron Light Research Institute (SLRI) on 2018 Apr 25
|
Crystal structure of the flavin reductase of Acinetobacter baumannii p-hydroxyphenylacetate 3-hydroxylase (HPAH) and identification of amino acid residues underlying its regulation by aromatic ligands.
Arch Biochem Biophys 653:24-38 (2018)
Yuenyao A, Petchyam N, Kamonsutthipaijit N, Chaiyen P, Pakotiprapha D
|
| RgGuinier |
2.7 |
nm |
| Dmax |
8.8 |
nm |
| VolumePorod |
92 |
nm3 |
|
|
UniProt ID: Q6Q271 (None-None) p-hydroxyphenylacetate 3-hydroxylase (HPAH), reductase component E251A mutant
|
|
|
|
| Sample: |
P-hydroxyphenylacetate 3-hydroxylase (HPAH), reductase component E251A mutant dimer, 71 kDa Acinetobacter baumannii protein
|
| Buffer: |
50 mM MOPS, 0.5 mM EDTA, 1 mM DTT, 50 mM NaCl, 1 mM HPA, 10 % glycerol, pH: 7 |
| Experiment: |
SAXS
data collected at BL1.3W, Synchrotron Light Research Institute (SLRI) on 2018 Apr 25
|
Crystal structure of the flavin reductase of Acinetobacter baumannii p-hydroxyphenylacetate 3-hydroxylase (HPAH) and identification of amino acid residues underlying its regulation by aromatic ligands.
Arch Biochem Biophys 653:24-38 (2018)
Yuenyao A, Petchyam N, Kamonsutthipaijit N, Chaiyen P, Pakotiprapha D
|
| RgGuinier |
2.8 |
nm |
| Dmax |
8.9 |
nm |
| VolumePorod |
86 |
nm3 |
|
|
UniProt ID: Q6Q271 (None-None) p-hydroxyphenylacetate 3-hydroxylase (HPAH), reductase component E251A mutant
|
|
|
|
| Sample: |
P-hydroxyphenylacetate 3-hydroxylase (HPAH), reductase component E251A mutant dimer, 71 kDa Acinetobacter baumannii protein
|
| Buffer: |
50 mM MOPS, 0.5 mM EDTA, 1 mM DTT, 50 mM NaCl, 1 mM HPA, 10 % glycerol, pH: 7 |
| Experiment: |
SAXS
data collected at BL1.3W, Synchrotron Light Research Institute (SLRI) on 2018 Apr 25
|
Crystal structure of the flavin reductase of Acinetobacter baumannii p-hydroxyphenylacetate 3-hydroxylase (HPAH) and identification of amino acid residues underlying its regulation by aromatic ligands.
Arch Biochem Biophys 653:24-38 (2018)
Yuenyao A, Petchyam N, Kamonsutthipaijit N, Chaiyen P, Pakotiprapha D
|
| RgGuinier |
2.9 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
82 |
nm3 |
|
|
UniProt ID: Q6Q271 (None-None) p-hydroxyphenylacetate 3-hydroxylase (HPAH), reductase component E251A mutant
|
|
|
|
| Sample: |
P-hydroxyphenylacetate 3-hydroxylase (HPAH), reductase component E251A mutant dimer, 71 kDa Acinetobacter baumannii protein
|
| Buffer: |
50 mM MOPS, 0.5 mM EDTA, 1 mM DTT, 50 mM NaCl, 1 mM HPA, 10 % glycerol, pH: 7 |
| Experiment: |
SAXS
data collected at BL1.3W, Synchrotron Light Research Institute (SLRI) on 2018 Apr 25
|
Crystal structure of the flavin reductase of Acinetobacter baumannii p-hydroxyphenylacetate 3-hydroxylase (HPAH) and identification of amino acid residues underlying its regulation by aromatic ligands.
Arch Biochem Biophys 653:24-38 (2018)
Yuenyao A, Petchyam N, Kamonsutthipaijit N, Chaiyen P, Pakotiprapha D
|
| RgGuinier |
2.9 |
nm |
| Dmax |
9.3 |
nm |
| VolumePorod |
89 |
nm3 |
|
|
UniProt ID: P02649 (19-317) Apolipoprotein E3
|
|
|
|
| Sample: |
Apolipoprotein E3 tetramer, 139 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 300 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Nov 29
|
The molecular basis for Apolipoprotein E4 as the major risk factor for late onset Alzheimer's disease.
J Mol Biol (2019)
Raulin AC, Kraft L, Al-Hilaly YK, Xue WF, McGeehan JE, Atack JR, Serpell L
|
| RgGuinier |
5.7 |
nm |
| Dmax |
19.5 |
nm |
| VolumePorod |
410 |
nm3 |
|
|
UniProt ID: P02649 (19-317) Apolipoprotein E4
|
|
|
|
| Sample: |
Apolipoprotein E4 tetramer, 139 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 300 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Nov 29
|
The molecular basis for Apolipoprotein E4 as the major risk factor for late onset Alzheimer's disease.
J Mol Biol (2019)
Raulin AC, Kraft L, Al-Hilaly YK, Xue WF, McGeehan JE, Atack JR, Serpell L
|
| RgGuinier |
5.8 |
nm |
| Dmax |
19.6 |
nm |
| VolumePorod |
430 |
nm3 |
|
|
UniProt ID: P25060 (261-382) Type II secretion system protein L, periplasmic domain
|
|
|
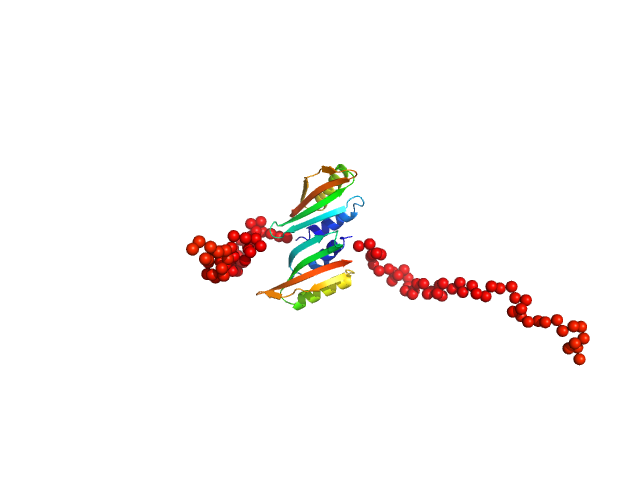
|
| Sample: |
Type II secretion system protein L, periplasmic domain dimer, 28 kDa Pseudomonas aeruginosa protein
|
| Buffer: |
50 mM TRIS, 100 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2016 Apr 8
|
Structure and oligomerization of the periplasmic domain of GspL from the type II secretion system of Pseudomonas aeruginosa.
Sci Rep 8(1):16760 (2018)
Fulara A, Vandenberghe I, Read RJ, Devreese B, Savvides SN
|
| RgGuinier |
2.2 |
nm |
| Dmax |
7.5 |
nm |
|
|
UniProt ID: P25060 (261-382) Type II secretion system protein L, periplasmic domain
|
|
|
|
| Sample: |
Type II secretion system protein L, periplasmic domain dimer, 28 kDa Pseudomonas aeruginosa protein
|
| Buffer: |
50 mM TRIS, 100 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2016 Apr 8
|
Structure and oligomerization of the periplasmic domain of GspL from the type II secretion system of Pseudomonas aeruginosa.
Sci Rep 8(1):16760 (2018)
Fulara A, Vandenberghe I, Read RJ, Devreese B, Savvides SN
|
| RgGuinier |
3.2 |
nm |
| Dmax |
10.5 |
nm |
|
|
UniProt ID: P16144-2 (1436-1666) Integrin beta-4 fragment with final part of the connecting segment and the third and fourth FnIII domains
|
|
|
|
| Sample: |
Integrin beta-4 fragment with final part of the connecting segment and the third and fourth FnIII domains monomer, 26 kDa Homo sapiens protein
|
| Buffer: |
20 mM Sodium Phosphate 150 mM NaCl 5% glycerol 3 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Nov 26
|
Integrin α6β4 Recognition of a Linear Motif of Bullous Pemphigoid Antigen BP230 Controls Its Recruitment to Hemidesmosomes.
Structure 27(6):952-964.e6 (2019)
Manso JA, Gómez-Hernández M, Carabias A, Alonso-García N, García-Rubio I, Kreft M, Sonnenberg A, de Pereda JM
|
| RgGuinier |
2.4 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
37 |
nm3 |
|
|