UniProt ID: P24300 (None-None) Xylose isomerase
|
|
|

|
| Sample: |
Xylose isomerase tetramer, 172 kDa Streptomyces rubiginosus protein
|
| Buffer: |
PBS, 50% Glycerol, 0.076 M NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2012 Apr 24
|
Standard proteins
Erica Valentini
|
| RgGuinier |
3.4 |
nm |
| Dmax |
9.7 |
nm |
| VolumePorod |
293 |
nm3 |
|
|
UniProt ID: V9P0A9 (None-None) Bacterial chalcone isomerase
|
|
|
|
| Sample: |
Bacterial chalcone isomerase hexamer, 194 kDa Eubacterium ramulus protein
|
| Buffer: |
50 mM sodium phosphate, pH: 6.8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Sep 23
|
Structure and catalytic mechanism of the evolutionarily unique bacterial chalcone isomerase.
Acta Crystallogr D Biol Crystallogr 71(Pt 4):907-17 (2015)
Thomsen M, Tuukkanen A, Dickerhoff J, Palm GJ, Kratzat H, Svergun DI, Weisz K, Bornscheuer UT, Hinrichs W
|
| RgGuinier |
4.0 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
320 |
nm3 |
|
|
UniProt ID: V9P0A9 (None-None) Chalcone isomerase with Naringenin
|
|
|
|
| Sample: |
Chalcone isomerase with Naringenin hexamer, 194 kDa Eubacterium ramulus protein
|
| Buffer: |
50 mM sodium phosphate, pH: 6.8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Sep 23
|
Structure and catalytic mechanism of the evolutionarily unique bacterial chalcone isomerase.
Acta Crystallogr D Biol Crystallogr 71(Pt 4):907-17 (2015)
Thomsen M, Tuukkanen A, Dickerhoff J, Palm GJ, Kratzat H, Svergun DI, Weisz K, Bornscheuer UT, Hinrichs W
|
| RgGuinier |
3.7 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
320 |
nm3 |
|
|
UniProt ID: V9P0A9 (None-None) Chalcone isomerase deltaLid
|
|
|
|
| Sample: |
Chalcone isomerase deltaLid hexamer, 181 kDa Eubacterium ramulus protein
|
| Buffer: |
50 mM sodium phosphate, pH: 6.8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Sep 23
|
Structure and catalytic mechanism of the evolutionarily unique bacterial chalcone isomerase.
Acta Crystallogr D Biol Crystallogr 71(Pt 4):907-17 (2015)
Thomsen M, Tuukkanen A, Dickerhoff J, Palm GJ, Kratzat H, Svergun DI, Weisz K, Bornscheuer UT, Hinrichs W
|
| RgGuinier |
3.6 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
270 |
nm3 |
|
|
UniProt ID: Q15149 (None-None) Plectin
|
|
|
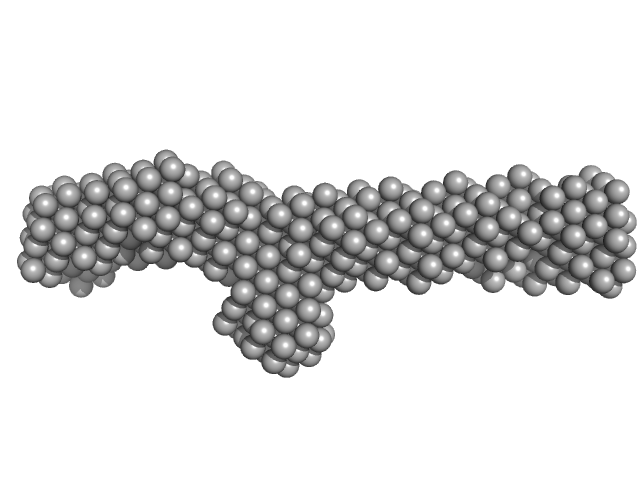
|
| Sample: |
Plectin monomer, 43 kDa Homo sapiens protein
|
| Buffer: |
20 mM Sodium Phosphate 150 mM NaCl 5% glycerol 2.5 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at cSAXS, Swiss Light Source on 2009 Jul 24
|
The structure of the plakin domain of plectin reveals a non-canonical SH3 domain interacting with its fourth spectrin repeat.
J Biol Chem 286(14):12429-38 (2011)
Ortega E, Buey RM, Sonnenberg A, de Pereda JM
|
| RgGuinier |
4.4 |
nm |
| Dmax |
14.5 |
nm |
| VolumePorod |
57 |
nm3 |
|
|
UniProt ID: P16144 (None-None) Integrin beta-4
|
|
|

|
| Sample: |
Integrin beta-4 monomer, 23 kDa Homo sapiens protein
|
| Buffer: |
20 mM Sodium Phosphate 150 mM NaCl 5% glycerol 3 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Aug 6
|
Combination of X-ray crystallography, SAXS and DEER to obtain the structure of the FnIII-3,4 domains of integrin α6β4.
Acta Crystallogr D Biol Crystallogr 71(Pt 4):969-85 (2015)
Alonso-García N, García-Rubio I, Manso JA, Buey RM, Urien H, Sonnenberg A, Jeschke G, de Pereda JM
|
| RgGuinier |
2.2 |
nm |
| Dmax |
7.0 |
nm |
| VolumePorod |
32 |
nm3 |
|
|
UniProt ID: P0A6B7 (None-None) Cysteine desulfurase IscS
|
|
|
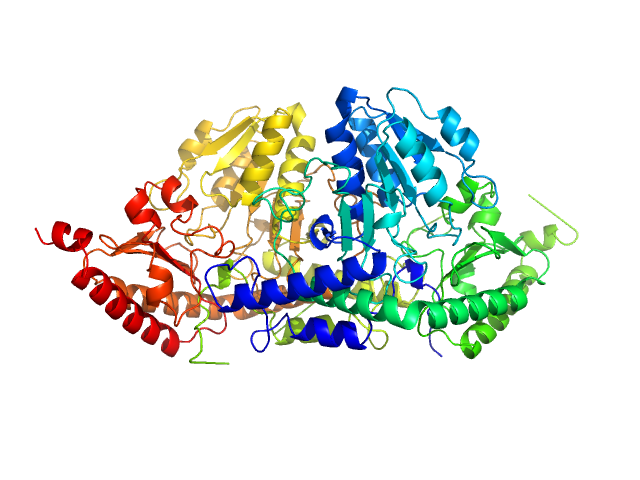
|
| Sample: |
Cysteine desulfurase IscS dimer, 87 kDa Escherichia coli protein
|
| Buffer: |
20 mM Tris-HCl 150 mM NaCl 10 mM β-mercaptoethanol, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Nov 5
|
Structural bases for the interaction of frataxin with the central components of iron-sulphur cluster assembly.
Nat Commun 1:95 (2010)
Prischi F, Konarev PV, Iannuzzi C, Pastore C, Adinolfi S, Martin SR, Svergun DI, Pastore A
|
| RgGuinier |
3.2 |
nm |
| Dmax |
10.9 |
nm |
|
|
UniProt ID: P0ACD4 (None-None) Iron-sulfur cluster assembly scaffold protein IscU
|
|
|

|
| Sample: |
Iron-sulfur cluster assembly scaffold protein IscU monomer, 14 kDa Escherichia coli protein
|
| Buffer: |
20 mM Tris-HCl 150 mM NaCl 10 mM β-mercaptoethanol, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Nov 5
|
Structural bases for the interaction of frataxin with the central components of iron-sulphur cluster assembly.
Nat Commun 1:95 (2010)
Prischi F, Konarev PV, Iannuzzi C, Pastore C, Adinolfi S, Martin SR, Svergun DI, Pastore A
|
| RgGuinier |
1.9 |
nm |
| Dmax |
6.4 |
nm |
|
|
UniProt ID: P27838 (None-None) Protein CyaY
|
|
|
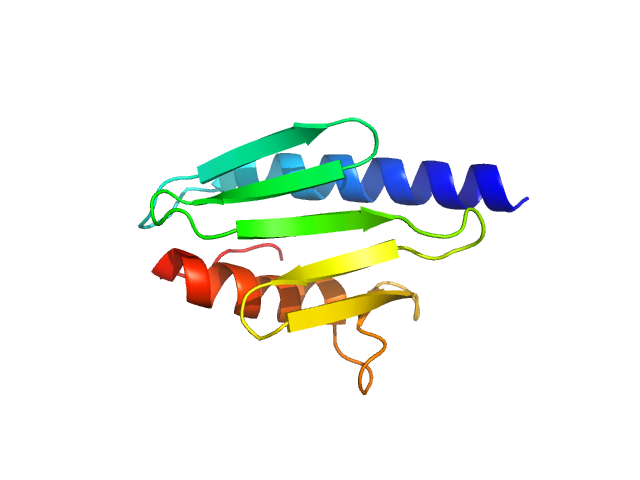
|
| Sample: |
Protein CyaY monomer, 12 kDa Escherichia coli protein
|
| Buffer: |
20 mM Tris-HCl 150 mM NaCl 10 mM β-mercaptoethanol, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Nov 5
|
Structural bases for the interaction of frataxin with the central components of iron-sulphur cluster assembly.
Nat Commun 1:95 (2010)
Prischi F, Konarev PV, Iannuzzi C, Pastore C, Adinolfi S, Martin SR, Svergun DI, Pastore A
|
| RgGuinier |
1.5 |
nm |
| Dmax |
5.0 |
nm |
|
|
UniProt ID: P0A6B7 (None-None) Cysteine desulfurase IscS
UniProt ID: P0ACD4 (None-None) Iron-sulfur cluster assembly scaffold protein IscU
|
|
|

|
| Sample: |
Cysteine desulfurase IscS dimer, 87 kDa Escherichia coli protein
Iron-sulfur cluster assembly scaffold protein IscU dimer, 29 kDa Escherichia coli protein
|
| Buffer: |
20 mM Tris-HCl 150 mM NaCl 10 mM β-mercaptoethanol, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Nov 5
|
Structural bases for the interaction of frataxin with the central components of iron-sulphur cluster assembly.
Nat Commun 1:95 (2010)
Prischi F, Konarev PV, Iannuzzi C, Pastore C, Adinolfi S, Martin SR, Svergun DI, Pastore A
|
| RgGuinier |
3.6 |
nm |
| Dmax |
12.1 |
nm |
|
|