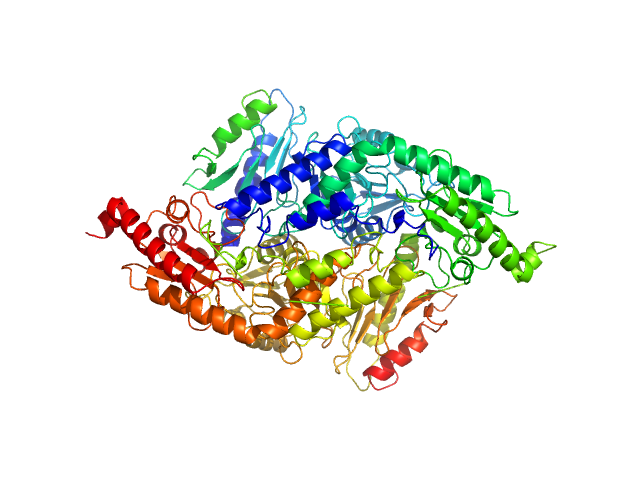
UniProt ID: P0A6B7 (None-None) Cysteine desulfurase IscS
UniProt ID: P27838 (None-None) Protein CyaY
|
|
|
|
| Sample: |
Cysteine desulfurase IscS dimer, 87 kDa Escherichia coli protein
Protein CyaY dimer, 24 kDa Escherichia coli protein
|
| Buffer: |
20 mM Tris-HCl, 50 mM NaCl, 10 mM β-mercaptoethanol and ferrous ammonium sulphate, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Nov 5
|
Structural bases for the interaction of frataxin with the central components of iron-sulphur cluster assembly.
Nat Commun 1:95 (2010)
Prischi F, Konarev PV, Iannuzzi C, Pastore C, Adinolfi S, Martin SR, Svergun DI, Pastore A
|
| RgGuinier |
3.1 |
nm |
| Dmax |
10.9 |
nm |
|
|
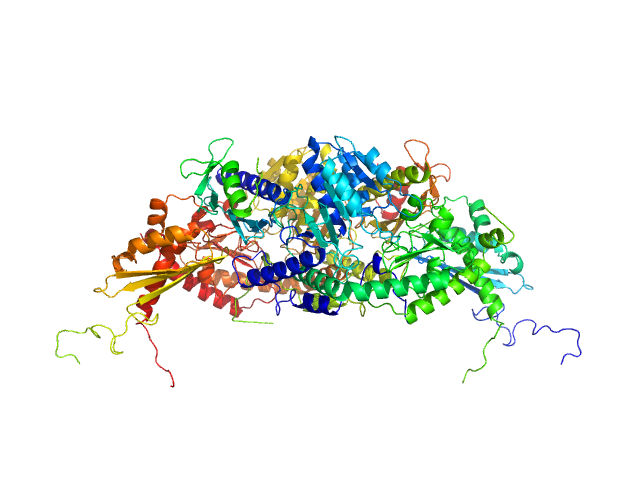
UniProt ID: P0A6B7 (None-None) Cysteine desulfurase IscS
UniProt ID: P0ACD4 (None-None) Iron-sulfur cluster assembly scaffold protein IscU
UniProt ID: P27838 (None-None) Protein CyaY
|
|
|
|
| Sample: |
Cysteine desulfurase IscS dimer, 87 kDa Escherichia coli protein
Iron-sulfur cluster assembly scaffold protein IscU dimer, 29 kDa Escherichia coli protein
Protein CyaY dimer, 24 kDa Escherichia coli protein
|
| Buffer: |
20 mM Tris-HCl 150 mM NaCl 10 mM β-mercaptoethanol, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Nov 5
|
Structural bases for the interaction of frataxin with the central components of iron-sulphur cluster assembly.
Nat Commun 1:95 (2010)
Prischi F, Konarev PV, Iannuzzi C, Pastore C, Adinolfi S, Martin SR, Svergun DI, Pastore A
|
| RgGuinier |
3.5 |
nm |
| Dmax |
11.9 |
nm |
|
|
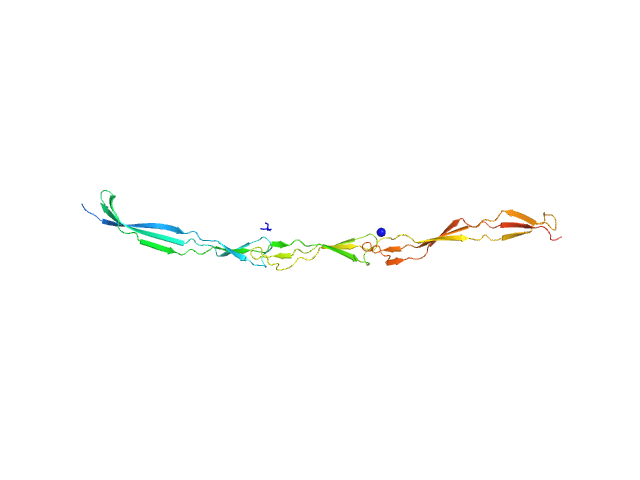
UniProt ID: Q2G2B2 (420-629) Surface protein G
|
|
|
|
| Sample: |
Surface protein G monomer, 24 kDa Staphylococcus aureus protein
|
| Buffer: |
20 mM Tris 200 mM NaCl 1 mM EDTA 20 mM Tris.Cl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Nov 12
|
Cooperative folding of intrinsically disordered domains drives assembly of a strong elongated protein.
Nat Commun 6:7271 (2015)
Gruszka DT, Whelan F, Farrance OE, Fung HK, Paci E, Jeffries CM, Svergun DI, Baldock C, Baumann CG, Brockwell DJ, Potts JR, Clarke J
|
| RgGuinier |
4.7 |
nm |
| Dmax |
19.0 |
nm |
| VolumePorod |
29 |
nm3 |
|
|
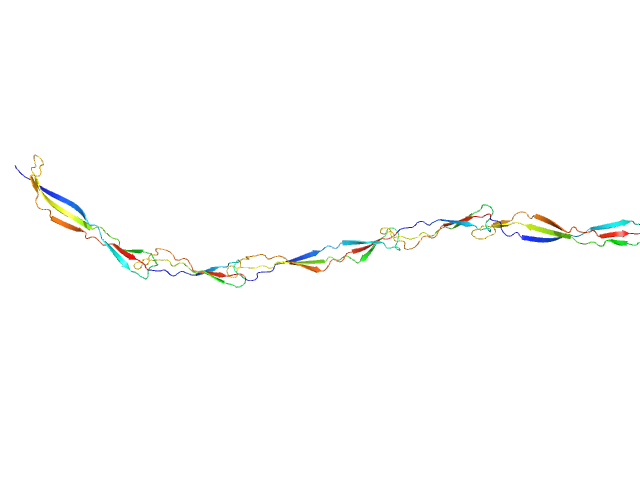
UniProt ID: Q2G2B2 (419-757) Surface protein G
|
|
|
|
| Sample: |
Surface protein G monomer, 39 kDa Staphylococcus aureus protein
|
| Buffer: |
20 mM Tris 200 mM NaCl 1 mM EDTA 20 mM Tris.Cl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Nov 12
|
Cooperative folding of intrinsically disordered domains drives assembly of a strong elongated protein.
Nat Commun 6:7271 (2015)
Gruszka DT, Whelan F, Farrance OE, Fung HK, Paci E, Jeffries CM, Svergun DI, Baldock C, Baumann CG, Brockwell DJ, Potts JR, Clarke J
|
| RgGuinier |
7.7 |
nm |
| Dmax |
30.5 |
nm |
| VolumePorod |
49 |
nm3 |
|
|
UniProt ID: Q2G2B2 (419-885) Surface protein G
|
|
|

|
| Sample: |
Surface protein G monomer, 53 kDa Staphylococcus aureus protein
|
| Buffer: |
20 mM Tris 200 mM NaCl 1 mM EDTA 20 mM Tris.Cl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Nov 12
|
Cooperative folding of intrinsically disordered domains drives assembly of a strong elongated protein.
Nat Commun 6:7271 (2015)
Gruszka DT, Whelan F, Farrance OE, Fung HK, Paci E, Jeffries CM, Svergun DI, Baldock C, Baumann CG, Brockwell DJ, Potts JR, Clarke J
|
| RgGuinier |
9.7 |
nm |
| Dmax |
38.5 |
nm |
| VolumePorod |
58 |
nm3 |
|
|
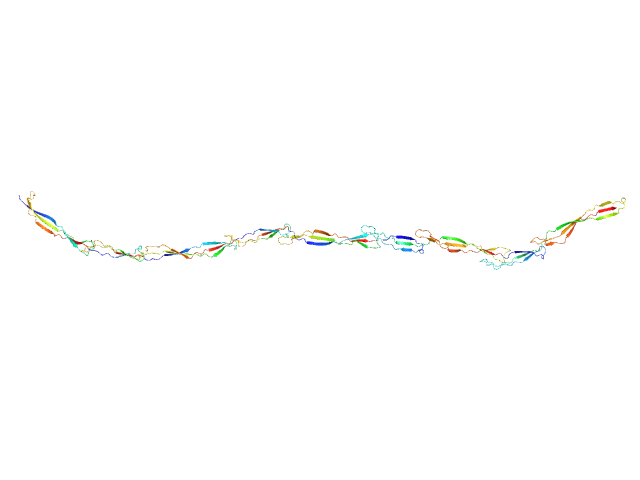
UniProt ID: Q2G2B2 (419-1013) Surface protein G
|
|
|
|
| Sample: |
Surface protein G monomer, 67 kDa Staphylococcus aureus protein
|
| Buffer: |
20 mM Tris 200 mM NaCl 1 mM EDTA 20 mM Tris.Cl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Nov 12
|
Cooperative folding of intrinsically disordered domains drives assembly of a strong elongated protein.
Nat Commun 6:7271 (2015)
Gruszka DT, Whelan F, Farrance OE, Fung HK, Paci E, Jeffries CM, Svergun DI, Baldock C, Baumann CG, Brockwell DJ, Potts JR, Clarke J
|
| RgGuinier |
12.0 |
nm |
| Dmax |
48.0 |
nm |
| VolumePorod |
87 |
nm3 |
|
|
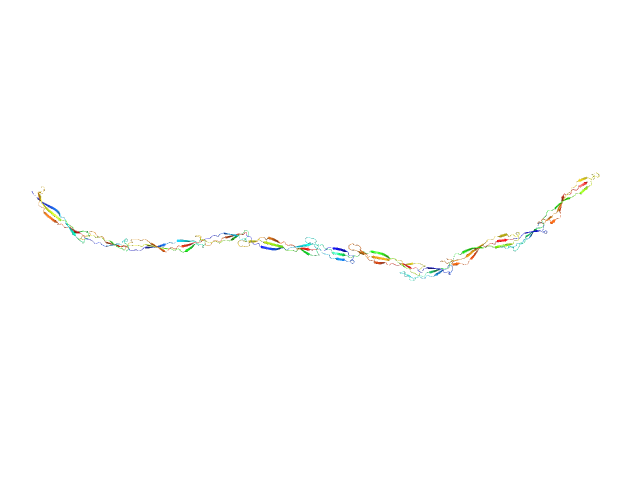
UniProt ID: Q2G2B2 (419-1141) Surface protein G
|
|
|
|
| Sample: |
Surface protein G monomer, 81 kDa Staphylococcus aureus protein
|
| Buffer: |
20 mM Tris 200 mM NaCl 1 mM EDTA 20 mM Tris.Cl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Nov 12
|
Cooperative folding of intrinsically disordered domains drives assembly of a strong elongated protein.
Nat Commun 6:7271 (2015)
Gruszka DT, Whelan F, Farrance OE, Fung HK, Paci E, Jeffries CM, Svergun DI, Baldock C, Baumann CG, Brockwell DJ, Potts JR, Clarke J
|
| RgGuinier |
14.1 |
nm |
| Dmax |
57.0 |
nm |
| VolumePorod |
89 |
nm3 |
|
|
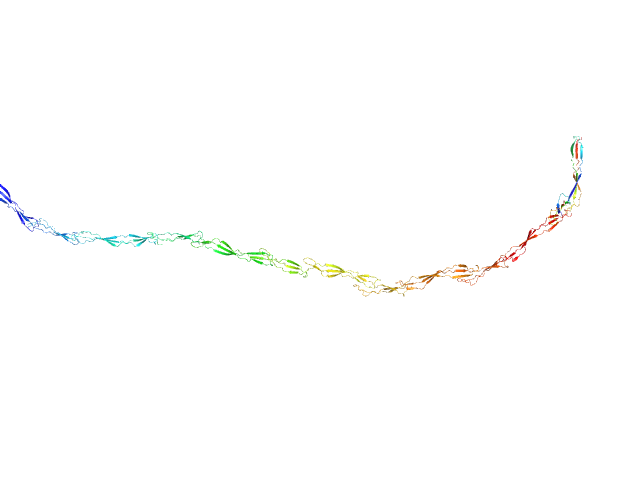
UniProt ID: Q2G2B2 (419-1269) Surface protein G
|
|
|
|
| Sample: |
Surface protein G monomer, 95 kDa Staphylococcus aureus protein
|
| Buffer: |
20 mM Tris 200 mM NaCl 1 mM EDTA 20 mM Tris.Cl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Nov 12
|
Cooperative folding of intrinsically disordered domains drives assembly of a strong elongated protein.
Nat Commun 6:7271 (2015)
Gruszka DT, Whelan F, Farrance OE, Fung HK, Paci E, Jeffries CM, Svergun DI, Baldock C, Baumann CG, Brockwell DJ, Potts JR, Clarke J
|
| RgGuinier |
15.9 |
nm |
| Dmax |
63.0 |
nm |
| VolumePorod |
122 |
nm3 |
|
|
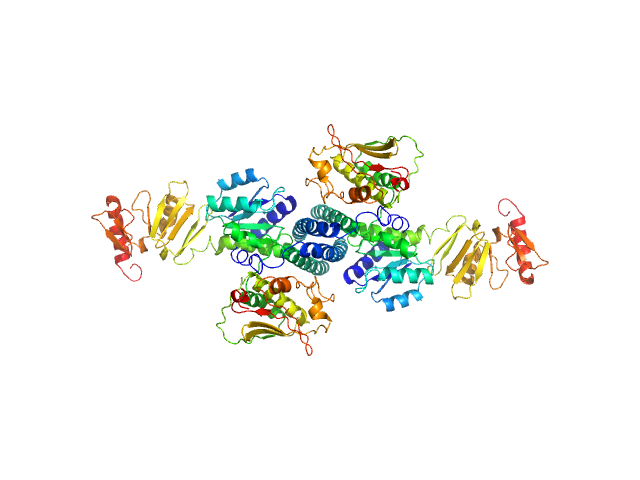
UniProt ID: Q8DMW4 (216-441) Histidine protein kinase
UniProt ID: Q8DMW5 (1-250) Response regulator
|
|
|
|
| Sample: |
Histidine protein kinase dimer, 54 kDa Streptococcus pneumoniae protein
Response regulator dimer, 61 kDa Streptococcus pneumoniae protein
|
| Buffer: |
20 mM Tris 200 mM NaCl 5% (v/v) Glycerol 5 mM β-mercaptoethanol, pH: 7.5 |
| Experiment: |
SAXS
data collected at Bruker Nanostar, IBBMC on 2012 May 16
|
Modeling the ComD/ComE/comcde interaction network using small angle X-ray scattering.
FEBS J 282(8):1538-53 (2015)
Sanchez D, Boudes M, van Tilbeurgh H, Durand D, Quevillon-Cheruel S
|
| RgGuinier |
4.0 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
175 |
nm3 |
|
|
UniProt ID: (None-None) comcde
UniProt ID: Q8DMW5 (1-250) Response regulator
|
|
|
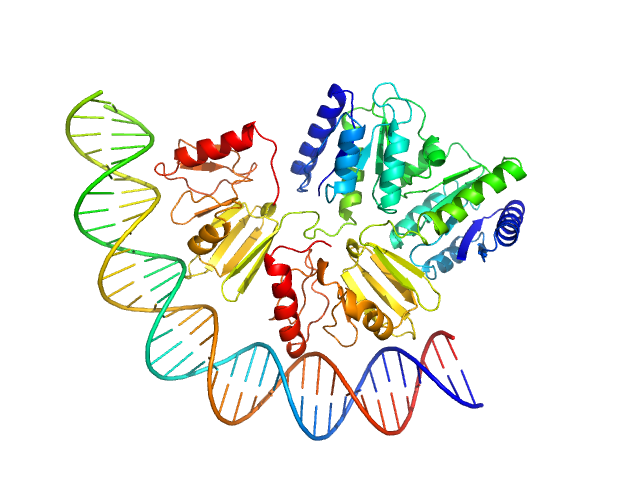
|
| Sample: |
Comcde, 24 kDa Streptococcus pneumoniae DNA
Response regulator dimer, 61 kDa Streptococcus pneumoniae protein
|
| Buffer: |
50 mM MES 500 mM NaCl 5% (v/v) Glycerol 5 mM β-mercaptoethanol, pH: 6.2 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2011 Feb 11
|
Modeling the ComD/ComE/comcde interaction network using small angle X-ray scattering.
FEBS J 282(8):1538-53 (2015)
Sanchez D, Boudes M, van Tilbeurgh H, Durand D, Quevillon-Cheruel S
|
| RgGuinier |
3.4 |
nm |
| Dmax |
10.7 |
nm |
| VolumePorod |
122 |
nm3 |
|
|