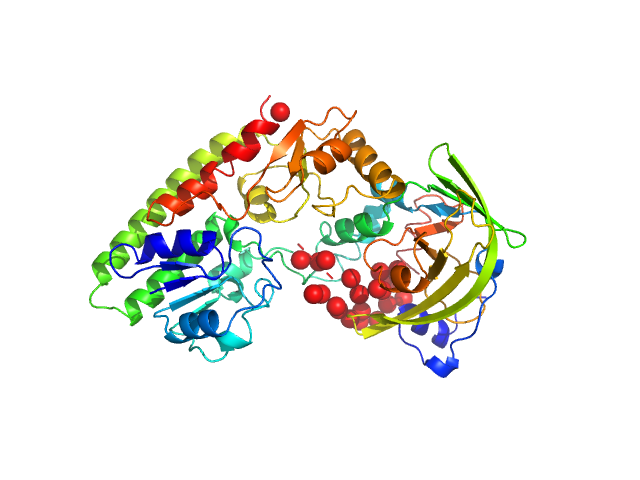
UniProt ID: V2JZU2 (None-None) High-affinity zinc transporter periplasmic component
UniProt ID: L6R5I5 (None-None) Zinc/cadmium-binding protein
|
|
|
|
| Sample: |
High-affinity zinc transporter periplasmic component monomer, 33 kDa Salmonella enterica subsp. … protein
Zinc/cadmium-binding protein monomer, 23 kDa Salmonella enterica subsp. … protein
|
| Buffer: |
50 mM HEPES 50 mM KCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2013 May 9
|
The Salmonella enterica ZinT structure, zinc affinity and interaction with the high-affinity uptake protein ZnuA provide insight into the management of periplasmic zinc.
Biochim Biophys Acta 1840(1):535-44 (2014)
Ilari A, Alaleona F, Tria G, Petrarca P, Battistoni A, Zamparelli C, Verzili D, Falconi M, Chiancone E
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
55 |
nm3 |
|
|
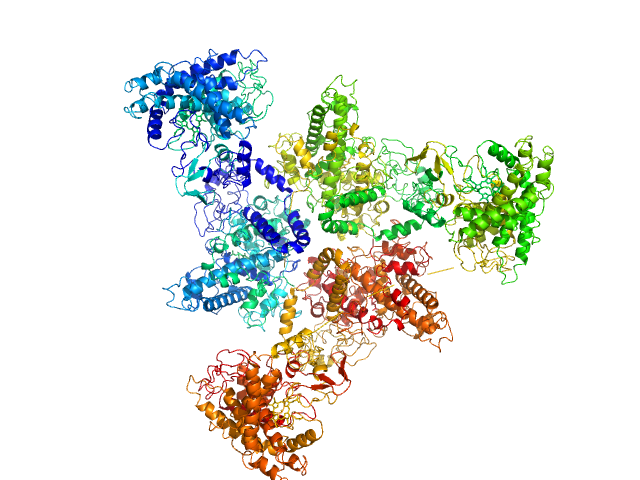
UniProt ID: Q6RET3 (None-None) Psi-producing oxygenase A
|
|
|
|
| Sample: |
Psi-producing oxygenase A trimer, 362 kDa Aspergillus nidulans protein
|
| Buffer: |
20 Mm HEPES, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Jun 24
|
A structural model of PpoA derived from SAXS-analysis-implications for substrate conversion.
Biochim Biophys Acta 1831(9):1449-57 (2013)
Koch C, Tria G, Fielding AJ, Brodhun F, Valerius O, Feussner K, Braus GH, Svergun DI, Bennati M, Feussner I
|
| RgGuinier |
5.4 |
nm |
| Dmax |
16.5 |
nm |
| VolumePorod |
540 |
nm3 |
|
|
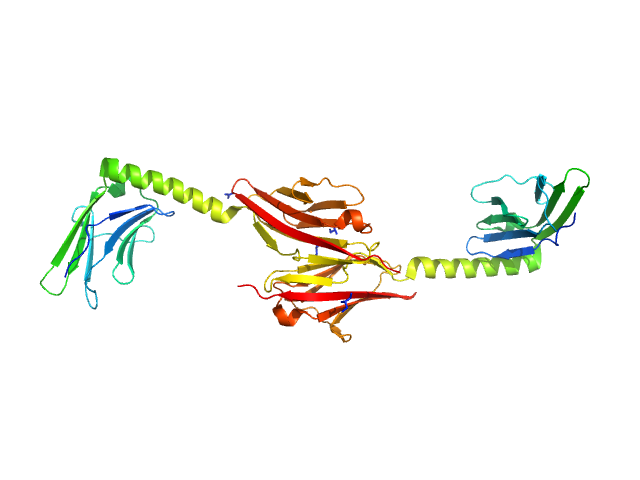
UniProt ID: P52179 (1225-1443) Myomesin-1
|
|
|
|
| Sample: |
Myomesin-1 dimer, 46 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris/HCl 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2005 May 7
|
Molecular basis of the C-terminal tail-to-tail assembly of the sarcomeric filament protein myomesin.
EMBO J 27(1):253-64 (2008)
Pinotsis N, Lange S, Perriard JC, Svergun DI, Wilmanns M
|
| RgGuinier |
4.0 |
nm |
| Dmax |
14.5 |
nm |
| VolumePorod |
56 |
nm3 |
|
|
UniProt ID: O95631 (None-None) Netrin-1
|
|
|

|
| Sample: |
Netrin-1 monomer, 49 kDa Homo sapiens protein
|
| Buffer: |
25 MES mM 200 mM NaCl 50 mM Tris 0.2 M ammonium sulfate (NH4)2(SO4) 1mM calcium chloride CaCl2, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Aug 26
|
The crystal structure of netrin-1 in complex with DCC reveals the bifunctionality of netrin-1 as a guidance cue.
Neuron 83(4):839-849 (2014)
Finci LI, Krüger N, Sun X, Zhang J, Chegkazi M, Wu Y, Schenk G, Mertens HDT, Svergun DI, Zhang Y, Wang JH, Meijers R
|
| RgGuinier |
3.9 |
nm |
| Dmax |
13.5 |
nm |
| VolumePorod |
100 |
nm3 |
|
|
UniProt ID: O95631 (None-None) Netrin-1
UniProt ID: (None-None) Deleted in Colorectal Cancer (FN5 & FN6)
|
|
|
|
| Sample: |
Netrin-1 monomer, 49 kDa Homo sapiens protein
Deleted in Colorectal Cancer (FN5 & FN6) monomer, 26 kDa Homo sapiens protein
|
| Buffer: |
25 MES mM 200 mM NaCl 50 mM Tris 0.2 M ammonium sulfate (NH4)2(SO4) 1mM calcium chloride CaCl2, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Oct 31
|
The crystal structure of netrin-1 in complex with DCC reveals the bifunctionality of netrin-1 as a guidance cue.
Neuron 83(4):839-849 (2014)
Finci LI, Krüger N, Sun X, Zhang J, Chegkazi M, Wu Y, Schenk G, Mertens HDT, Svergun DI, Zhang Y, Wang JH, Meijers R
|
| RgGuinier |
5.1 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
120 |
nm3 |
|
|
UniProt ID: O95631 (None-None) Netrin-1
UniProt ID: (None-None) Deleted in Colorectal Cancer (FN5 & FN6) M933R mutant
|
|
|
|
| Sample: |
Netrin-1 monomer, 49 kDa Homo sapiens protein
Deleted in Colorectal Cancer (FN5 & FN6) M933R mutant monomer, 26 kDa Homo sapiens protein
|
| Buffer: |
25 MES mM 200 mM NaCl 50 mM Tris 0.2 M ammonium sulfate (NH4)2(SO4) 1mM calcium chloride CaCl2, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Aug 26
|
The crystal structure of netrin-1 in complex with DCC reveals the bifunctionality of netrin-1 as a guidance cue.
Neuron 83(4):839-849 (2014)
Finci LI, Krüger N, Sun X, Zhang J, Chegkazi M, Wu Y, Schenk G, Mertens HDT, Svergun DI, Zhang Y, Wang JH, Meijers R
|
| RgGuinier |
4.0 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
95 |
nm3 |
|
|
UniProt ID: P00698 (19-147) Lysozyme C
|
|
|

|
| Sample: |
Lysozyme C monomer, 14 kDa Gallus gallus protein
|
| Buffer: |
20 mM Sodium Acetate/HEPES, pH: 6.8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2013 Feb 17
|
Correlation Map, a goodness-of-fit test for one-dimensional X-ray scattering spectra.
Nat Methods 12(5):419-22 (2015)
Franke D, Jeffries CM, Svergun DI
|
| RgGuinier |
1.5 |
nm |
| Dmax |
4.8 |
nm |
| VolumePorod |
24 |
nm3 |
|
|
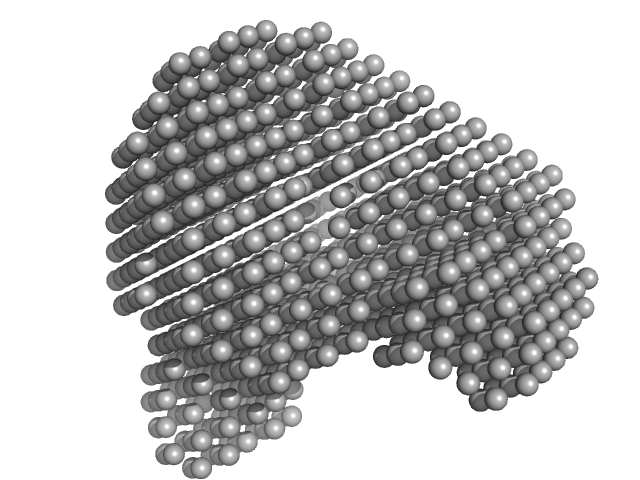
UniProt ID: P02768 (25-609) Human serum albumin monomer
|
|
|
|
| Sample: |
Human serum albumin monomer monomer, 66 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2014 Jan 22
|
Correlation Map, a goodness-of-fit test for one-dimensional X-ray scattering spectra.
Nat Methods 12(5):419-22 (2015)
Franke D, Jeffries CM, Svergun DI
|
| RgGuinier |
2.8 |
nm |
| Dmax |
8.4 |
nm |
| VolumePorod |
103 |
nm3 |
|
|
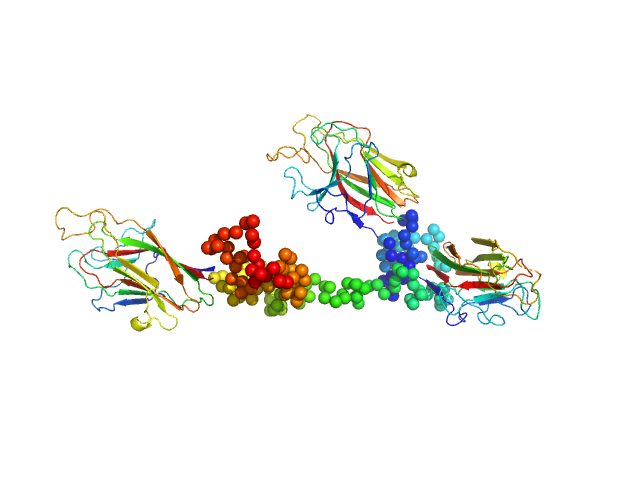
UniProt ID: Q51817 (982-1662) K1K2K3 adhesin modules of lysine-specific (Kgp) gingipain
|
|
|
|
| Sample: |
K1K2K3 adhesin modules of lysine-specific (Kgp) gingipain monomer, 74 kDa Porphyromonas gingivalis W83 protein
|
| Buffer: |
10 mM TRIS 150 mM NaCl, pH: 7.6 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSess, University of Sydney on 2010 Aug 15
|
The modular structure of haemagglutinin/adhesin regions in gingipains of Porphyromonas gingivalis.
Mol Microbiol 81(5):1358-73 (2011)
Li N, Yun P, Jeffries CM, Langley D, Gamsjaeger R, Church WB, Hunter N, Collyer CA
|
| RgGuinier |
4.3 |
nm |
| Dmax |
14.7 |
nm |
|
|
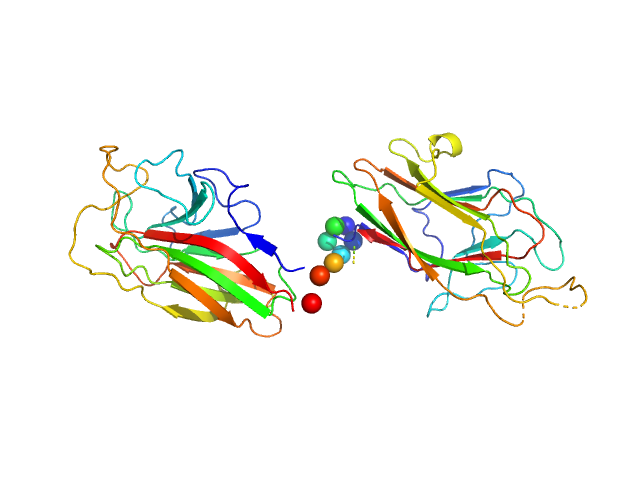
UniProt ID: Q51817 (982-1334) K1K2 adhesin modules of lysine-specific (Kgp) gingipain
|
|
|
|
| Sample: |
K1K2 adhesin modules of lysine-specific (Kgp) gingipain monomer, 38 kDa Porphyromonas gingivalis W83 protein
|
| Buffer: |
10 mM TRIS 150 mM NaCl, pH: 7.6 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSess, University of Sydney on 2010 Aug 15
|
The modular structure of haemagglutinin/adhesin regions in gingipains of Porphyromonas gingivalis.
Mol Microbiol 81(5):1358-73 (2011)
Li N, Yun P, Jeffries CM, Langley D, Gamsjaeger R, Church WB, Hunter N, Collyer CA
|
| RgGuinier |
2.9 |
nm |
| Dmax |
9.5 |
nm |
|
|