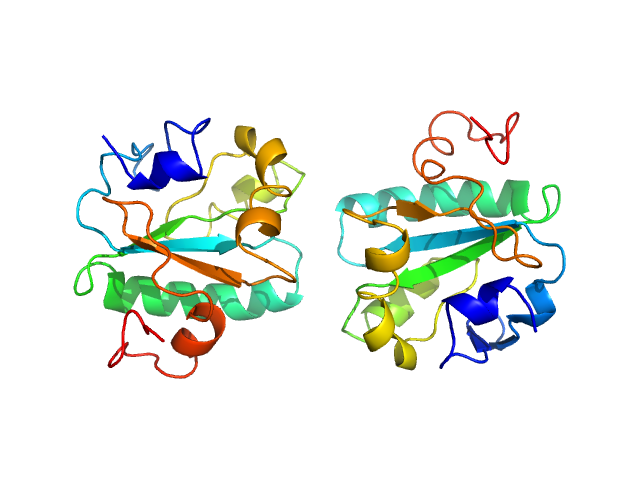
UniProt ID: O77404 (None-None) Tryparedoxin K102E
|
|
|
|
| Sample: |
Tryparedoxin K102E monomer, 16 kDa Trypanosoma brucei brucei protein
|
| Buffer: |
10 mM HEPES pH 7.5, 50 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 May 31
|
Inhibitor-induced dimerization of an essential oxidoreductase from African Trypanosomes.
Angew Chem Int Ed Engl (2019)
Wagner A, Le TA, Brennich M, Klein P, Bader N, Diehl E, Paszek D, Weickhmann AK, Dirdjaja N, Krauth-Siegel RL, Engels B, Opatz T, Schindelin H, Hellmich UA
|
| RgGuinier |
1.6 |
nm |
| Dmax |
6.0 |
nm |
| VolumePorod |
29 |
nm3 |
|
|
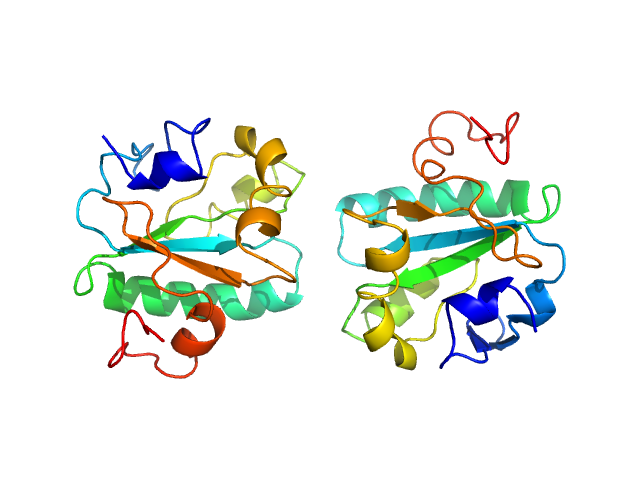
UniProt ID: O77404 (None-None) Tryparedoxin K102E
|
|
|
|
| Sample: |
Tryparedoxin K102E monomer, 16 kDa Trypanosoma brucei brucei protein
|
| Buffer: |
10 mM HEPES pH 7.5, 50 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 May 31
|
Inhibitor-induced dimerization of an essential oxidoreductase from African Trypanosomes.
Angew Chem Int Ed Engl (2019)
Wagner A, Le TA, Brennich M, Klein P, Bader N, Diehl E, Paszek D, Weickhmann AK, Dirdjaja N, Krauth-Siegel RL, Engels B, Opatz T, Schindelin H, Hellmich UA
|
| RgGuinier |
1.6 |
nm |
| Dmax |
4.3 |
nm |
| VolumePorod |
26 |
nm3 |
|
|
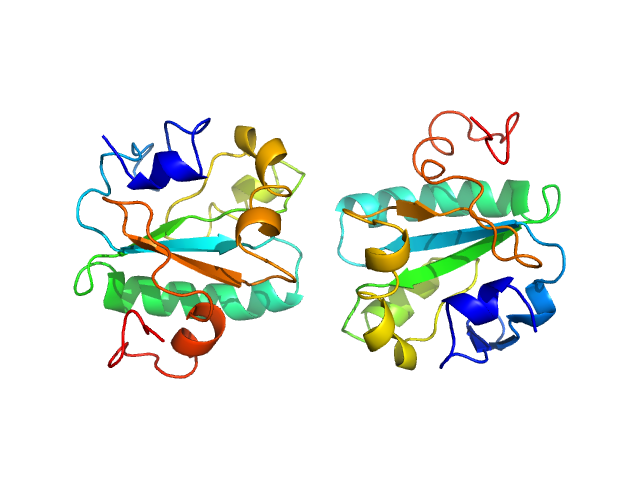
UniProt ID: O77404 (None-None) Tryparedoxin K102E
|
|
|
|
| Sample: |
Tryparedoxin K102E, 16 kDa Trypanosoma brucei brucei protein
|
| Buffer: |
10 mM HEPES pH 7.5, 50 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 May 31
|
Inhibitor-induced dimerization of an essential oxidoreductase from African Trypanosomes.
Angew Chem Int Ed Engl (2019)
Wagner A, Le TA, Brennich M, Klein P, Bader N, Diehl E, Paszek D, Weickhmann AK, Dirdjaja N, Krauth-Siegel RL, Engels B, Opatz T, Schindelin H, Hellmich UA
|
| RgGuinier |
1.8 |
nm |
| Dmax |
6.2 |
nm |
| VolumePorod |
37 |
nm3 |
|
|
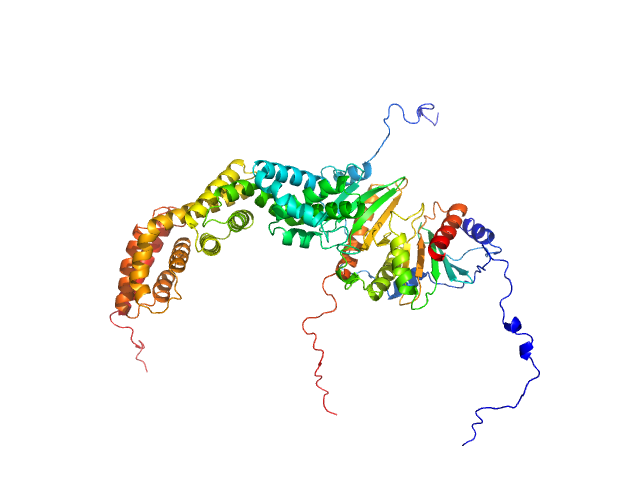
UniProt ID: D4HVP1 (None-None) Type III secretion protein HrpG
UniProt ID: D4HVP4 (None-None) Type III secretion protein HrpV
UniProt ID: D4HVM8 (None-None) Hypersensitivity response secretion protein HrpJ
|
|
|
|
| Sample: |
Type III secretion protein HrpG, 19 kDa Erwinia amylovora protein
Type III secretion protein HrpV, 13 kDa Erwinia amylovora protein
Hypersensitivity response secretion protein HrpJ, 42 kDa Erwinia amylovora protein
|
| Buffer: |
20mM Tris-Cl, pH 8.0, 100mM NaCl, 2mM DTT, 0.5mM EDTA, pH: 8 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2016 Apr 3
|
Migration of Type III Secretion System Transcriptional Regulators Links Gene Expression to Secretion.
MBio 9(4) (2018)
Charova SN, Gazi AD, Mylonas E, Pozidis C, Sabarit B, Anagnostou D, Psatha K, Aivaliotis M, Beuzon CR, Panopoulos NJ, Kokkinidis M
|
| RgGuinier |
4.0 |
nm |
| Dmax |
16.0 |
nm |
|
|
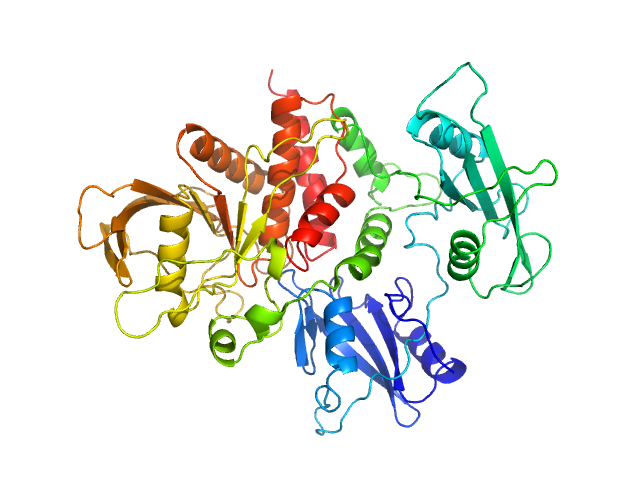
UniProt ID: Q06124-2 (1-529) Tyrosine-protein phosphatase non-receptor type 11
|
|
|
|
| Sample: |
Tyrosine-protein phosphatase non-receptor type 11 monomer, 61 kDa Homo sapiens protein
|
| Buffer: |
50 mM ADA, 2 mM TCEP, pH: 6.5 |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2016 Mar 28
|
Mechanism of activating mutations and allosteric drug inhibition of the phosphatase SHP2.
Nat Commun 9(1):4507 (2018)
Pádua RAP, Sun Y, Marko I, Pitsawong W, Stiller JB, Otten R, Kern D
|
| RgGuinier |
2.7 |
nm |
| Dmax |
8.9 |
nm |
|
|
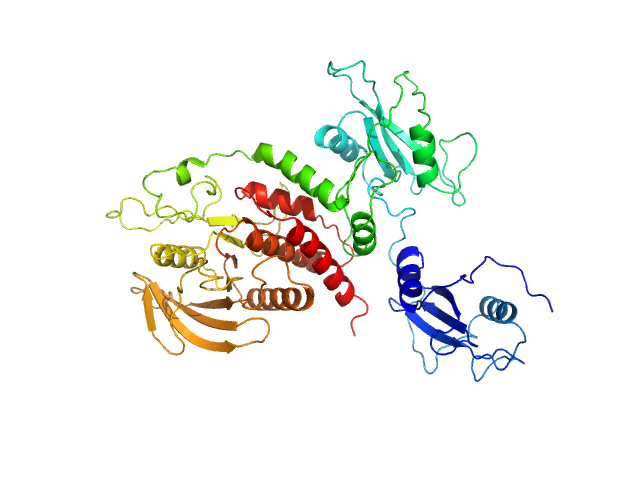
UniProt ID: Q06124-2 (1-529) Tyrosine-protein phosphatase non-receptor type 11 E76K
|
|
|
|
| Sample: |
Tyrosine-protein phosphatase non-receptor type 11 E76K monomer, 61 kDa Homo sapiens protein
|
| Buffer: |
50 mM ADA, 2 mM TCEP, pH: 6.5 |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2016 Mar 28
|
Mechanism of activating mutations and allosteric drug inhibition of the phosphatase SHP2.
Nat Commun 9(1):4507 (2018)
Pádua RAP, Sun Y, Marko I, Pitsawong W, Stiller JB, Otten R, Kern D
|
| RgGuinier |
2.9 |
nm |
| Dmax |
9.5 |
nm |
|
|
UniProt ID: None (None-None) Membrane scaffold protein 1D1 (deuterated, 75%)
UniProt ID: None (None-None) 1-palmitoyl-2-palmitoleoyl-sn-glycero-3-phosphocholine (deuteration: 78% head, 92% acyl)
UniProt ID: Q9LF79 (None-None) Calcium-transporting ATPase 8, plasma membrane-type
|
|
|
|
| Sample: |
Membrane scaffold protein 1D1 (deuterated, 75%) dimer, 49 kDa protein
1-palmitoyl-2-palmitoleoyl-sn-glycero-3-phosphocholine (deuteration: 78% head, 92% acyl), 1 kDa Escherichia coli
Calcium-transporting ATPase 8, plasma membrane-type monomer, 118 kDa Arabidopsis thaliana protein
|
| Buffer: |
30 mM Tris, 150 mM NaCl, 1mM MgCl2, 1 mM CaCl2, pH: 7.5 |
| Experiment: |
SANS
data collected at SANS-1, Heinz Maier-Leibnitz Zentrum on 2017 Aug 28
|
Structural basis for activation of plasma-membrane Ca2+-ATPase by calmodulin.
Commun Biol 1:206 (2018)
Nitsche J, Josts I, Heidemann J, Mertens HD, Maric S, Moulin M, Haertlein M, Busch S, Forsyth VT, Svergun DI, Uetrecht C, Tidow H
|
| RgGuinier |
4.0 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
202 |
nm3 |
|
|
UniProt ID: None (None-None) Membrane scaffold protein 1D1 (deuterated, 75%)
UniProt ID: None (None-None) 1-palmitoyl-2-palmitoleoyl-sn-glycero-3-phosphocholine (deuteration: 78% head, 92% acyl)
UniProt ID: Q9LF79 (None-None) Calcium-transporting ATPase 8, plasma membrane-type
UniProt ID: P59220 (None-None) Calmodulin-7 (deuterated 75%)
|
|
|
|
| Sample: |
Membrane scaffold protein 1D1 (deuterated, 75%) dimer, 49 kDa protein
1-palmitoyl-2-palmitoleoyl-sn-glycero-3-phosphocholine (deuteration: 78% head, 92% acyl), 1 kDa Escherichia coli
Calcium-transporting ATPase 8, plasma membrane-type monomer, 118 kDa Arabidopsis thaliana protein
Calmodulin-7 (deuterated 75%) monomer, Arabidopsis thaliana protein
|
| Buffer: |
30 mM Tris, 150 mM NaCl, 1mM MgCl2, 1 mM CaCl2, pH: 7.5 |
| Experiment: |
SANS
data collected at SANS-1, Heinz Maier-Leibnitz Zentrum on 2017 Aug 28
|
Structural basis for activation of plasma-membrane Ca2+-ATPase by calmodulin.
Commun Biol 1:206 (2018)
Nitsche J, Josts I, Heidemann J, Mertens HD, Maric S, Moulin M, Haertlein M, Busch S, Forsyth VT, Svergun DI, Uetrecht C, Tidow H
|
| RgGuinier |
4.3 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
217 |
nm3 |
|
|
UniProt ID: None (None-None) Membrane scaffold protein 1D1 (deuterated, 75%)
UniProt ID: None (None-None) 1-palmitoyl-2-palmitoleoyl-sn-glycero-3-phosphocholine (deuteration: 78% head, 92% acyl)
UniProt ID: Q9LF79 (None-None) Calcium-transporting ATPase 8, plasma membrane-type
UniProt ID: P59220 (None-None) Calmodulin-7
|
|
|
|
| Sample: |
Membrane scaffold protein 1D1 (deuterated, 75%) dimer, 49 kDa protein
1-palmitoyl-2-palmitoleoyl-sn-glycero-3-phosphocholine (deuteration: 78% head, 92% acyl), 1 kDa Escherichia coli
Calcium-transporting ATPase 8, plasma membrane-type monomer, 118 kDa Arabidopsis thaliana protein
Calmodulin-7 monomer, Arabidopsis thaliana protein
|
| Buffer: |
30 mM Tris, 150 mM NaCl, 1mM MgCl2, 1 mM CaCl2, pH: 7.5 |
| Experiment: |
SANS
data collected at SANS-1, Heinz Maier-Leibnitz Zentrum on 2017 Aug 28
|
Structural basis for activation of plasma-membrane Ca2+-ATPase by calmodulin.
Commun Biol 1:206 (2018)
Nitsche J, Josts I, Heidemann J, Mertens HD, Maric S, Moulin M, Haertlein M, Busch S, Forsyth VT, Svergun DI, Uetrecht C, Tidow H
|
| RgGuinier |
4.8 |
nm |
| Dmax |
18.0 |
nm |
| VolumePorod |
297 |
nm3 |
|
|
UniProt ID: None (None-None) Membrane scaffold protein 1D1 (deuterated, 75%)
UniProt ID: None (None-None) 1-palmitoyl-2-palmitoleoyl-sn-glycero-3-phosphocholine (deuteration: 78% head, 92% acyl)
UniProt ID: Q9LF79 (None-None) Calcium-transporting ATPase 8, plasma membrane-type
|
|
|
|
| Sample: |
Membrane scaffold protein 1D1 (deuterated, 75%) dimer, 49 kDa protein
1-palmitoyl-2-palmitoleoyl-sn-glycero-3-phosphocholine (deuteration: 78% head, 92% acyl), 1 kDa Escherichia coli
Calcium-transporting ATPase 8, plasma membrane-type monomer, 118 kDa Arabidopsis thaliana protein
|
| Buffer: |
30 mM Tris, 150 mM NaCl, 1mM MgCl2, 1 mM CaCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Sep 8
|
Structural basis for activation of plasma-membrane Ca2+-ATPase by calmodulin.
Commun Biol 1:206 (2018)
Nitsche J, Josts I, Heidemann J, Mertens HD, Maric S, Moulin M, Haertlein M, Busch S, Forsyth VT, Svergun DI, Uetrecht C, Tidow H
|
| RgGuinier |
5.3 |
nm |
| Dmax |
20.0 |
nm |
| VolumePorod |
626 |
nm3 |
|
|