UniProt ID: P12978 (381-455) Epstein-Barr nuclear antigen 2
UniProt ID: Q15326-1 (480-602) Zinc finger MYND domain-containing protein 11
|
|
|

|
| Sample: |
Epstein-Barr nuclear antigen 2 dimer, 16 kDa Human gammaherpesvirus 4 protein
Zinc finger MYND domain-containing protein 11 tetramer, 60 kDa Homo sapiens protein
|
| Buffer: |
20mM Tris-HCl, 100mM NaCl, 2% Sucrose and 1mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Nov 29
|
Increased association between Epstein-Barr virus EBNA2 from type 2 strains and the transcriptional repressor BS69 restricts B cell growth
(2018)
Ponnusamy R, Khatri R, Correia P, Mancini E, Farrell P, West M
|
| RgGuinier |
4.6 |
nm |
| Dmax |
13.8 |
nm |
| VolumePorod |
72 |
nm3 |
|
|
UniProt ID: Q69022 (348-422) Epstein-Barr nuclear antigen 2
UniProt ID: Q15326-1 (480-602) Zinc finger MYND domain-containing protein 11
|
|
|

|
| Sample: |
Epstein-Barr nuclear antigen 2 dimer, 16 kDa Human gammaherpesvirus 4 protein
Zinc finger MYND domain-containing protein 11 hexamer, 90 kDa Homo sapiens protein
|
| Buffer: |
20mM Tris-HCl, 100mM NaCl, 2% Sucrose and 1mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Sep 23
|
Increased association between Epstein-Barr virus EBNA2 from type 2 strains and the transcriptional repressor BS69 restricts B cell growth
(2018)
Ponnusamy R, Khatri R, Correia P, Mancini E, Farrell P, West M
|
| RgGuinier |
4.7 |
nm |
| Dmax |
14.5 |
nm |
| VolumePorod |
239 |
nm3 |
|
|
UniProt ID: A0A1H7TQR5 (None-None) Glucosamine kinase
|
|
|
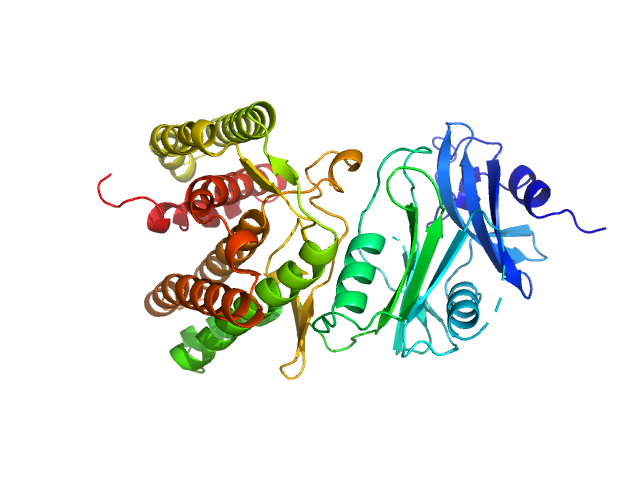
|
| Sample: |
Glucosamine kinase monomer, 48 kDa Streptacidiphilus jiangxiensis protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, 10 mM MgCl2, 5 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Nov 24
|
Molecular Fingerprints for a Novel Enzyme Family in Actinobacteria with Glucosamine Kinase Activity.
MBio 10(3) (2019)
Manso JA, Nunes-Costa D, Macedo-Ribeiro S, Empadinhas N, Pereira PJB
|
| RgGuinier |
2.6 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
72 |
nm3 |
|
|
UniProt ID: A0A1H7TQR5 (None-None) Glucosamine kinase
|
|
|
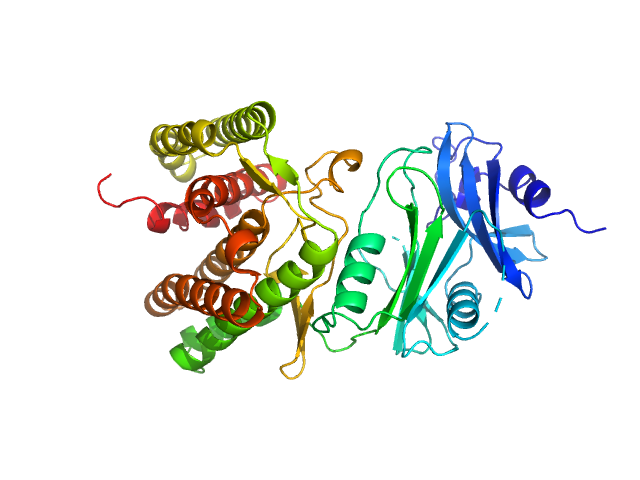
|
| Sample: |
Glucosamine kinase monomer, 48 kDa Streptacidiphilus jiangxiensis protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, 10 mM MgCl2, 5 mM DTT, 0.2 M D-glucosamine, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Nov 20
|
Molecular Fingerprints for a Novel Enzyme Family in Actinobacteria with Glucosamine Kinase Activity.
MBio 10(3) (2019)
Manso JA, Nunes-Costa D, Macedo-Ribeiro S, Empadinhas N, Pereira PJB
|
| RgGuinier |
2.6 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
78 |
nm3 |
|
|
UniProt ID: A0A1H7TQR5 (None-None) Glucosamine kinase
|
|
|
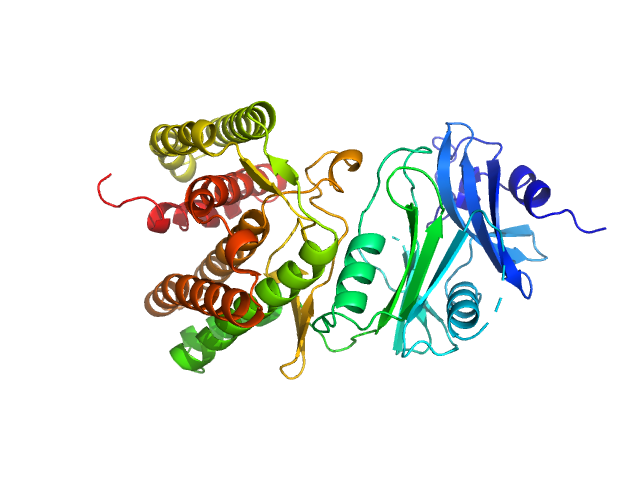
|
| Sample: |
Glucosamine kinase monomer, 48 kDa Streptacidiphilus jiangxiensis protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, 10 mM MgCl2, 5 mM DTT, 1 mM ATP, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Nov 24
|
Molecular Fingerprints for a Novel Enzyme Family in Actinobacteria with Glucosamine Kinase Activity.
MBio 10(3) (2019)
Manso JA, Nunes-Costa D, Macedo-Ribeiro S, Empadinhas N, Pereira PJB
|
| RgGuinier |
2.5 |
nm |
| Dmax |
7.8 |
nm |
| VolumePorod |
73 |
nm3 |
|
|
UniProt ID: A0A1H7TQR5 (None-None) Glucosamine kinase
|
|
|
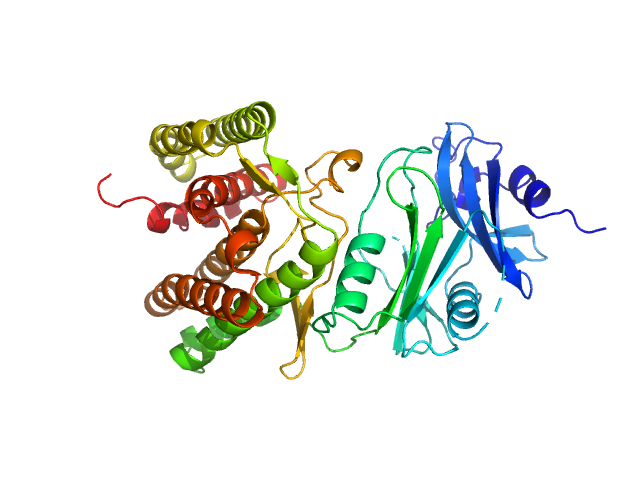
|
| Sample: |
Glucosamine kinase monomer, 48 kDa Streptacidiphilus jiangxiensis protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, 10 mM MgCl2, 5 mM DTT, 0.2 M D-glucosamine, 1 mM ATP, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Nov 20
|
Molecular Fingerprints for a Novel Enzyme Family in Actinobacteria with Glucosamine Kinase Activity.
MBio 10(3) (2019)
Manso JA, Nunes-Costa D, Macedo-Ribeiro S, Empadinhas N, Pereira PJB
|
| RgGuinier |
2.5 |
nm |
| Dmax |
7.7 |
nm |
| VolumePorod |
76 |
nm3 |
|
|
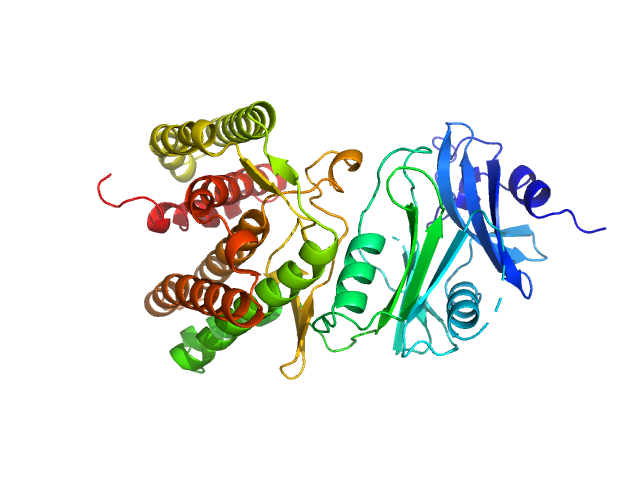
UniProt ID: A0A1H7TQR5 (None-None) Glucosamine kinase
|
|
|
|
| Sample: |
Glucosamine kinase monomer, 48 kDa Streptacidiphilus jiangxiensis protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, 10 mM MgCl2, 5 mM DTT, 50 mM D-glucose, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Apr 13
|
Molecular Fingerprints for a Novel Enzyme Family in Actinobacteria with Glucosamine Kinase Activity.
MBio 10(3) (2019)
Manso JA, Nunes-Costa D, Macedo-Ribeiro S, Empadinhas N, Pereira PJB
|
| RgGuinier |
2.6 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
73 |
nm3 |
|
|
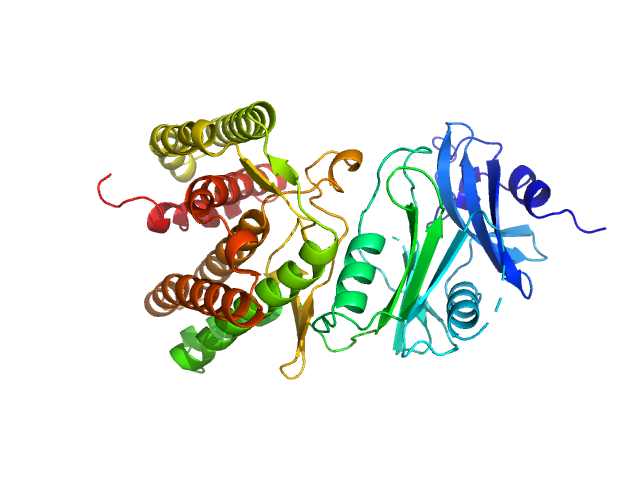
UniProt ID: A0A1H7TQR5 (None-None) Glucosamine kinase
|
|
|
|
| Sample: |
Glucosamine kinase monomer, 48 kDa Streptacidiphilus jiangxiensis protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, 10 mM MgCl2, 5 mM DTT, 50 mM D-glucose, 1 mM ATP, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Nov 24
|
Molecular Fingerprints for a Novel Enzyme Family in Actinobacteria with Glucosamine Kinase Activity.
MBio 10(3) (2019)
Manso JA, Nunes-Costa D, Macedo-Ribeiro S, Empadinhas N, Pereira PJB
|
| RgGuinier |
2.5 |
nm |
| Dmax |
7.7 |
nm |
| VolumePorod |
73 |
nm3 |
|
|
UniProt ID: Q8I374 (None-None) Proteasome activator PA28
|
|
|

|
| Sample: |
Proteasome activator PA28 heptamer, 232 kDa Plasmodium falciparum protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, 0.5 mM TCEP, 0.1% sodium azide, pH: 7.4 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2017 Dec 2
|
The structure of the PA28-20S proteasome complex from Plasmodium falciparum and implications for proteostasis.
Nat Microbiol 4(11):1990-2000 (2019)
Xie SC, Metcalfe RD, Hanssen E, Yang T, Gillett DL, Leis AP, Morton CJ, Kuiper MJ, Parker MW, Spillman NJ, Wong W, Tsu C, Dick LR, Griffin MDW, Tilley L
|
| RgGuinier |
4.3 |
nm |
| Dmax |
12.9 |
nm |
| VolumePorod |
484 |
nm3 |
|
|
UniProt ID: O60828 (1-192) Polyglutamine-binding protein 1 p.Lys192Serfs*7
|
|
|
|
| Sample: |
Polyglutamine-binding protein 1 p.Lys192Serfs*7 dimer, 47 kDa Homo sapiens protein
|
| Buffer: |
Phosphate-buffered saline, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2013 Feb 15
|
Frameshift PQBP-1 mutants K192Sfs*7 and R153Sfs*41 implicated in X-linked intellectual disability form stable dimers.
J Struct Biol (2019)
Rahman SK, Okazawa H, Chen YW
|
| RgGuinier |
3.5 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
114 |
nm3 |
|
|