UniProt ID: P53396-1 (None-None) ATP-citrate synthase
|
|
|
|
| Sample: |
ATP-citrate synthase tetramer, 458 kDa Homo sapiens protein
|
| Buffer: |
20mM HEPES, 150mM NaCl, 50mM Tris, 20mM citrate, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 May 6
|
Structure of ATP citrate lyase and the origin of citrate synthase in the Krebs cycle.
Nature 568(7753):571-575 (2019)
Verschueren KHG, Blanchet C, Felix J, Dansercoer A, De Vos D, Bloch Y, Van Beeumen J, Svergun D, Gutsche I, Savvides SN, Verstraete K
|
| RgGuinier |
6.1 |
nm |
| Dmax |
17.5 |
nm |
| VolumePorod |
747 |
nm3 |
|
|
UniProt ID: P53396-1 (None-None) ATP-citrate synthase
|
|
|
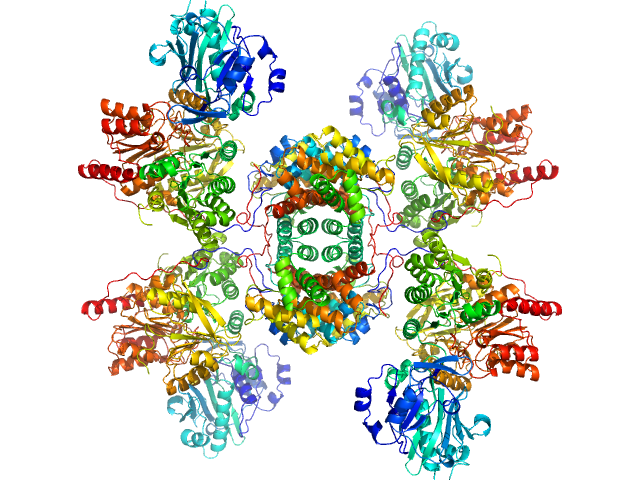
|
| Sample: |
ATP-citrate synthase tetramer, 458 kDa Homo sapiens protein
|
| Buffer: |
20mM HEPES, 150mM NaCl, 50mM Tris, 20mM citrate, 2mM CoA, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 May 5
|
Structure of ATP citrate lyase and the origin of citrate synthase in the Krebs cycle.
Nature 568(7753):571-575 (2019)
Verschueren KHG, Blanchet C, Felix J, Dansercoer A, De Vos D, Bloch Y, Van Beeumen J, Svergun D, Gutsche I, Savvides SN, Verstraete K
|
| RgGuinier |
5.8 |
nm |
| Dmax |
16.5 |
nm |
| VolumePorod |
709 |
nm3 |
|
|
UniProt ID: A0A2H4PMI3 (17-549) Aldehyde dehydrogenase 12
|
|
|
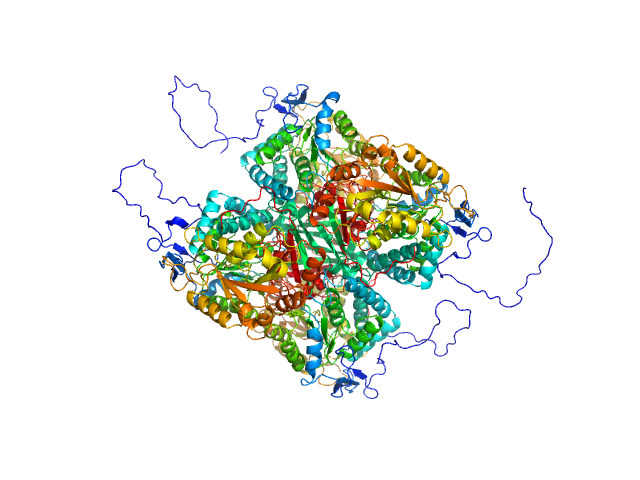
|
| Sample: |
Aldehyde dehydrogenase 12 tetramer, 242 kDa Zea mays protein
|
| Buffer: |
50 mM Tris-HCl, 50 mM NaCl, 0.5 mM TCEP, and 5% (v/v) glycerol, pH: 7.8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2016 Dec 6
|
Structural and Biochemical Characterization of Aldehyde Dehydrogenase 12, the Last Enzyme of Proline Catabolism in Plants.
J Mol Biol (2018)
Korasick DA, Končitíková R, Kopečná M, Hájková E, Vigouroux A, Moréra S, Becker DF, Šebela M, Tanner JJ, Kopečný D
|
| RgGuinier |
4.1 |
nm |
| Dmax |
14.4 |
nm |
| VolumePorod |
351 |
nm3 |
|
|
UniProt ID: A0A0H2XGG9 (None-None) N-acetylglucosamine-6-phosphate deacetylase
|
|
|
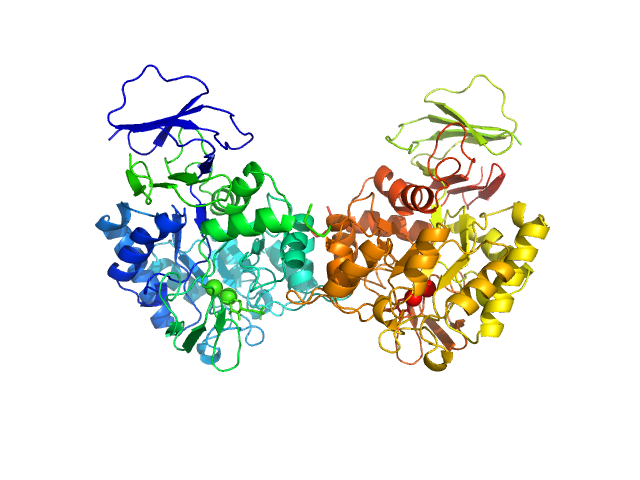
|
| Sample: |
N-acetylglucosamine-6-phosphate deacetylase dimer, 86 kDa Staphylococcus aureus protein
|
| Buffer: |
20 mM Tris-HCl 150 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2016 Apr 26
|
Functional and solution structure studies of amino sugar deacetylase and deaminase enzymes from Staphylococcus aureus.
FEBS Lett (2018)
Davies JS, Coombes D, Horne CR, Pearce FG, Friemann R, North RA, Dobson RCJ
|
| RgGuinier |
3.2 |
nm |
| Dmax |
10.1 |
nm |
| VolumePorod |
107 |
nm3 |
|
|
UniProt ID: A8YZR7 (None-None) Glucosamine-6-phosphate deaminase
|
|
|
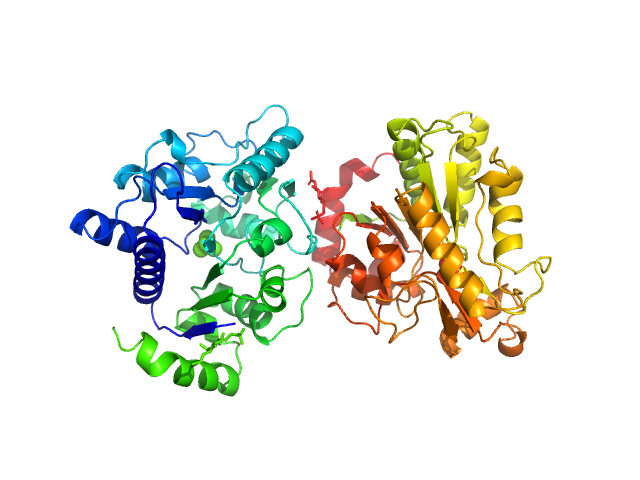
|
| Sample: |
Glucosamine-6-phosphate deaminase dimer, 57 kDa Staphylococcus aureus protein
|
| Buffer: |
20 mM Tris-HCl 150 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2016 Apr 26
|
Functional and solution structure studies of amino sugar deacetylase and deaminase enzymes from Staphylococcus aureus.
FEBS Lett (2018)
Davies JS, Coombes D, Horne CR, Pearce FG, Friemann R, North RA, Dobson RCJ
|
| RgGuinier |
2.7 |
nm |
| Dmax |
8.9 |
nm |
| VolumePorod |
84 |
nm3 |
|
|
UniProt ID: P15873 (None-None) Proliferating cell nuclear antigen
|
|
|

|
| Sample: |
Proliferating cell nuclear antigen trimer, 117 kDa Saccharomyces cerevisiae protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 5% glycerol, 1 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2018 Aug 6
|
Conformational Flexibility of Ubiquitin-Modified and SUMO-Modified PCNA Shown by Full-Ensemble Hybrid Methods.
J Mol Biol 430(24):5294-5303 (2018)
Powers KT, Lavering ED, Washington MT
|
| RgGuinier |
4.1 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
238 |
nm3 |
|
|
UniProt ID: P15873 (None-None) Proliferating cell nuclear antigen
|
|
|

|
| Sample: |
Proliferating cell nuclear antigen trimer, 125 kDa Saccharomyces cerevisiae protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 5% glycerol, 1 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2018 Aug 6
|
Conformational Flexibility of Ubiquitin-Modified and SUMO-Modified PCNA Shown by Full-Ensemble Hybrid Methods.
J Mol Biol 430(24):5294-5303 (2018)
Powers KT, Lavering ED, Washington MT
|
| RgGuinier |
4.7 |
nm |
| Dmax |
16.9 |
nm |
| VolumePorod |
310 |
nm3 |
|
|
UniProt ID: P12978 (381-455) Epstein-Barr nuclear antigen 2
|
|
|
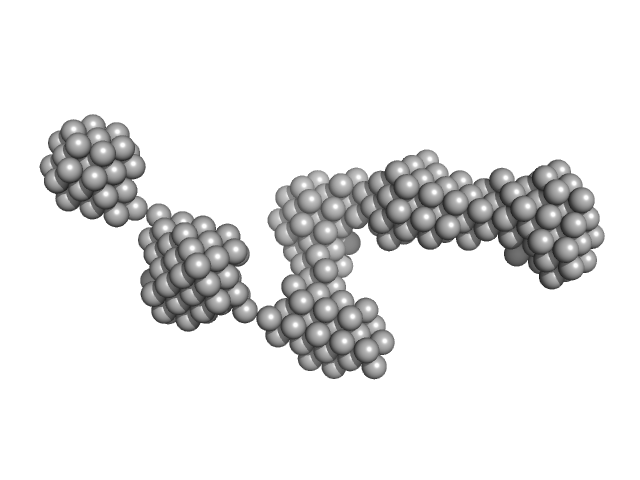
|
| Sample: |
Epstein-Barr nuclear antigen 2 dimer, 16 kDa Human gammaherpesvirus 4 protein
|
| Buffer: |
20mM Tris-HCl, 100mM NaCl, 2% Sucrose and 1mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Sep 23
|
Increased association between Epstein-Barr virus EBNA2 from type 2 strains and the transcriptional repressor BS69 restricts B cell growth
(2018)
Ponnusamy R, Khatri R, Correia P, Mancini E, Farrell P, West M
|
| RgGuinier |
2.7 |
nm |
| Dmax |
9.3 |
nm |
| VolumePorod |
19 |
nm3 |
|
|
UniProt ID: Q69022 (348-422) Epstein-Barr nuclear antigen 2
|
|
|
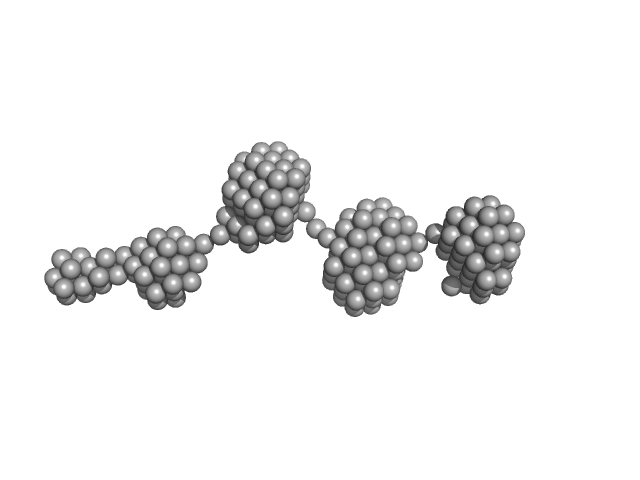
|
| Sample: |
Epstein-Barr nuclear antigen 2 dimer, 16 kDa Human gammaherpesvirus 4 protein
|
| Buffer: |
20mM Tris-HCl, 100mM NaCl, 2% Sucrose and 1mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Sep 23
|
Increased association between Epstein-Barr virus EBNA2 from type 2 strains and the transcriptional repressor BS69 restricts B cell growth
(2018)
Ponnusamy R, Khatri R, Correia P, Mancini E, Farrell P, West M
|
| RgGuinier |
2.8 |
nm |
| Dmax |
10.4 |
nm |
| VolumePorod |
20 |
nm3 |
|
|
UniProt ID: Q15326-1 (480-602) Zinc finger MYND domain-containing protein 11
|
|
|

|
| Sample: |
Zinc finger MYND domain-containing protein 11 tetramer, 60 kDa Homo sapiens protein
|
| Buffer: |
20mM Tris-HCl, 100mM NaCl, 2% Sucrose and 1mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Sep 23
|
Increased association between Epstein-Barr virus EBNA2 from type 2 strains and the transcriptional repressor BS69 restricts B cell growth
(2018)
Ponnusamy R, Khatri R, Correia P, Mancini E, Farrell P, West M
|
| RgGuinier |
5.3 |
nm |
| Dmax |
16.9 |
nm |
| VolumePorod |
83 |
nm3 |
|
|