UniProt ID: (None-None) 169 bp DNA (145 bp Widom 601, flanked by 12bp DNA)
UniProt ID: P06897 (None-None) Histone H2A type 1
UniProt ID: P02281 (None-None) Histone H2B 1.1
UniProt ID: P84233 (1-136) Histone H3.2
UniProt ID: P62799 (None-None) Histone H4
|
|
|
|
| Sample: |
169 bp DNA (145 bp Widom 601, flanked by 12bp DNA) monomer, 52 kDa DNA
Histone H2A type 1 monomer, 14 kDa Xenopus laevis protein
Histone H2B 1.1 monomer, 14 kDa Xenopus laevis protein
Histone H3.2 monomer, 15 kDa Xenopus laevis protein
Histone H4 monomer, 11 kDa Xenopus laevis protein
|
| Buffer: |
10 mM Tris, 100 mM NaCl, 2 mM MgCl2, 0.1 mM EDTA, 1 mM DTT, 60% (w/v) sucrose, ADP-BeF3 (0.5 mM ADP, 4 mM NaF, 0.6 mM BeCl2), pH: 7.8 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2015 Oct 24
|
The ATPase motor of the Chd1 chromatin remodeler stimulates DNA unwrapping from the nucleosome.
Nucleic Acids Res 46(10):4978-4990 (2018)
Tokuda JM, Ren R, Levendosky RF, Tay RJ, Yan M, Pollack L, Bowman GD
|
| RgGuinier |
4.8 |
nm |
| Dmax |
14.0 |
nm |
|
|
UniProt ID: P32657 (118-1274) Chromodomain-helicase-DNA-binding protein 1
UniProt ID: (None-None) 169 bp DNA (145 bp Widom 601, flanked by 12bp DNA)
UniProt ID: P06897 (None-None) Histone H2A type 1
UniProt ID: P02281 (None-None) Histone H2B 1.1
UniProt ID: P84233 (1-136) Histone H3.2
UniProt ID: P62799 (None-None) Histone H4
|
|
|
|
| Sample: |
Chromodomain-helicase-DNA-binding protein 1 dimer, 266 kDa Saccharomyces cerevisiae protein
169 bp DNA (145 bp Widom 601, flanked by 12bp DNA) monomer, 52 kDa DNA
Histone H2A type 1 monomer, 14 kDa Xenopus laevis protein
Histone H2B 1.1 monomer, 14 kDa Xenopus laevis protein
Histone H3.2 monomer, 15 kDa Xenopus laevis protein
Histone H4 monomer, 11 kDa Xenopus laevis protein
|
| Buffer: |
10 mM Tris, 100 mM NaCl, 2 mM MgCl2, 0.1 mM EDTA, 1 mM DTT, 60% (w/v) sucrose, pH: 7.8 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2015 Oct 24
|
The ATPase motor of the Chd1 chromatin remodeler stimulates DNA unwrapping from the nucleosome.
Nucleic Acids Res 46(10):4978-4990 (2018)
Tokuda JM, Ren R, Levendosky RF, Tay RJ, Yan M, Pollack L, Bowman GD
|
| RgGuinier |
5.2 |
nm |
| Dmax |
12.8 |
nm |
|
|
UniProt ID: P32657 (118-1274) Chromodomain-helicase-DNA-binding protein 1
UniProt ID: (None-None) 169 bp DNA (145 bp Widom 601, flanked by 12bp DNA)
UniProt ID: P06897 (None-None) Histone H2A type 1
UniProt ID: P02281 (None-None) Histone H2B 1.1
UniProt ID: P84233 (1-136) Histone H3.2
UniProt ID: P62799 (None-None) Histone H4
|
|
|
|
| Sample: |
Chromodomain-helicase-DNA-binding protein 1 dimer, 266 kDa Saccharomyces cerevisiae protein
169 bp DNA (145 bp Widom 601, flanked by 12bp DNA) monomer, 52 kDa DNA
Histone H2A type 1 monomer, 14 kDa Xenopus laevis protein
Histone H2B 1.1 monomer, 14 kDa Xenopus laevis protein
Histone H3.2 monomer, 15 kDa Xenopus laevis protein
Histone H4 monomer, 11 kDa Xenopus laevis protein
|
| Buffer: |
10 mM Tris, 100 mM NaCl, 2 mM MgCl2, 0.1 mM EDTA, 1 mM DTT, 60% (w/v) sucrose, ADP-BeF3 (0.5 mM ADP, 4 mM NaF, 0.6 mM BeCl2), pH: 7.8 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2015 Oct 24
|
The ATPase motor of the Chd1 chromatin remodeler stimulates DNA unwrapping from the nucleosome.
Nucleic Acids Res 46(10):4978-4990 (2018)
Tokuda JM, Ren R, Levendosky RF, Tay RJ, Yan M, Pollack L, Bowman GD
|
| RgGuinier |
5.3 |
nm |
| Dmax |
16.5 |
nm |
|
|
UniProt ID: P32657 (118-1274) Chromodomain-helicase-DNA-binding protein 1
UniProt ID: (None-None) 169 bp DNA (145 bp Widom 601, flanked by 12bp DNA)
UniProt ID: P06897 (None-None) Histone H2A type 1
UniProt ID: P02281 (None-None) Histone H2B 1.1
UniProt ID: P84233 (1-136) Histone H3.2
UniProt ID: P62799 (None-None) Histone H4
|
|
|
|
| Sample: |
Chromodomain-helicase-DNA-binding protein 1 dimer, 266 kDa Saccharomyces cerevisiae protein
169 bp DNA (145 bp Widom 601, flanked by 12bp DNA) monomer, 52 kDa DNA
Histone H2A type 1 monomer, 14 kDa Xenopus laevis protein
Histone H2B 1.1 monomer, 14 kDa Xenopus laevis protein
Histone H3.2 monomer, 15 kDa Xenopus laevis protein
Histone H4 monomer, 11 kDa Xenopus laevis protein
|
| Buffer: |
10 mM Tris, 100 mM NaCl, 2 mM MgCl2, 0.1 mM EDTA, 1 mM DTT, 60% (w/v) sucrose, 0.5 mM AMP-PNP, pH: 7.8 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2015 Oct 24
|
The ATPase motor of the Chd1 chromatin remodeler stimulates DNA unwrapping from the nucleosome.
Nucleic Acids Res 46(10):4978-4990 (2018)
Tokuda JM, Ren R, Levendosky RF, Tay RJ, Yan M, Pollack L, Bowman GD
|
| RgGuinier |
5.6 |
nm |
| Dmax |
16.7 |
nm |
|
|
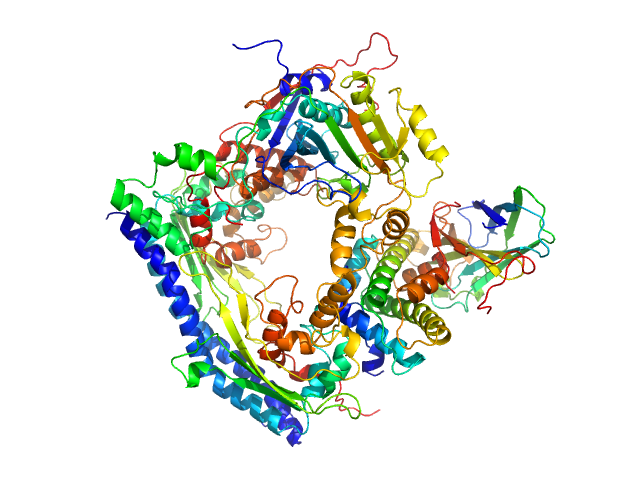
UniProt ID: P53853 (1-225) Vacuolar protein sorting-associated protein 75 (1-225 aa)
UniProt ID: Q07794 (1-436) Histone acetyltransferase RTT109
UniProt ID: P32447 (1-169) Histone chaperone ASF1
UniProt ID: P84233 (35-135) Histone H3.2 (35-135 aa)
UniProt ID: P62799 (1-103) Histone H4
|
|
|
|
| Sample: |
Vacuolar protein sorting-associated protein 75 (1-225 aa) dimer, 53 kDa Saccharomyces cerevisiae protein
Histone acetyltransferase RTT109 monomer, 50 kDa Saccharomyces cerevisiae protein
Histone chaperone ASF1 monomer, 19 kDa protein
Histone H3.2 (35-135 aa) monomer, 12 kDa Xenopus laevis protein
Histone H4 monomer, 11 kDa Xenopus laevis protein
|
| Buffer: |
50 mM citrate, 150 mM NaCl, 5 mM BME, 100% D2O, pH: 6.5 |
| Experiment: |
SANS
data collected at KWS1, FRM2 on 2017 Mar 3
|
Histone chaperone exploits intrinsic disorder to switch acetylation specificity.
Nat Commun 10(1):3435 (2019)
Danilenko N, Lercher L, Kirkpatrick J, Gabel F, Codutti L, Carlomagno T
|
| RgGuinier |
3.5 |
nm |
| Dmax |
11.8 |
nm |
|
|
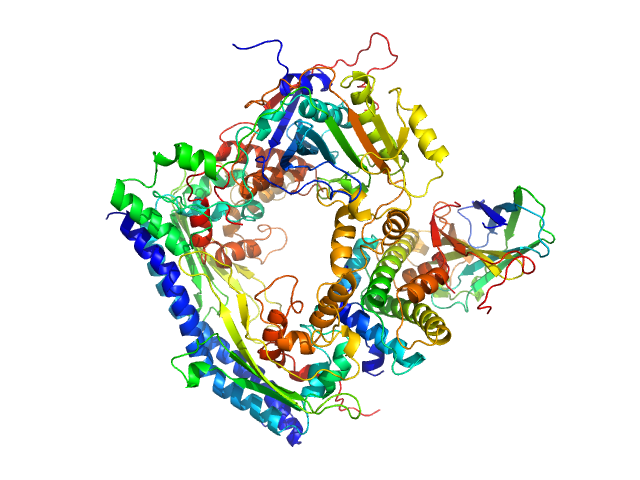
UniProt ID: P53853 (1-225) Vacuolar protein sorting-associated protein 75 (1-225 aa)
UniProt ID: Q07794 (1-436) Histone acetyltransferase RTT109
UniProt ID: P32447 (1-169) Histone chaperone ASF1
UniProt ID: P84233 (35-135) Histone H3.2 (35-135 aa)
UniProt ID: P62799 (1-103) Histone H4
|
|
|
|
| Sample: |
Vacuolar protein sorting-associated protein 75 (1-225 aa) dimer, 53 kDa Saccharomyces cerevisiae protein
Histone acetyltransferase RTT109 monomer, 50 kDa Saccharomyces cerevisiae protein
Histone chaperone ASF1 monomer, 19 kDa protein
Histone H3.2 (35-135 aa) monomer, 12 kDa Xenopus laevis protein
Histone H4 monomer, 11 kDa Xenopus laevis protein
|
| Buffer: |
50 mM citrate, 150 mM NaCl, 5 mM BME, 100% D2O, pH: 6.5 |
| Experiment: |
SANS
data collected at KWS1, FRM2 on 2017 Mar 4
|
Histone chaperone exploits intrinsic disorder to switch acetylation specificity.
Nat Commun 10(1):3435 (2019)
Danilenko N, Lercher L, Kirkpatrick J, Gabel F, Codutti L, Carlomagno T
|
|
|
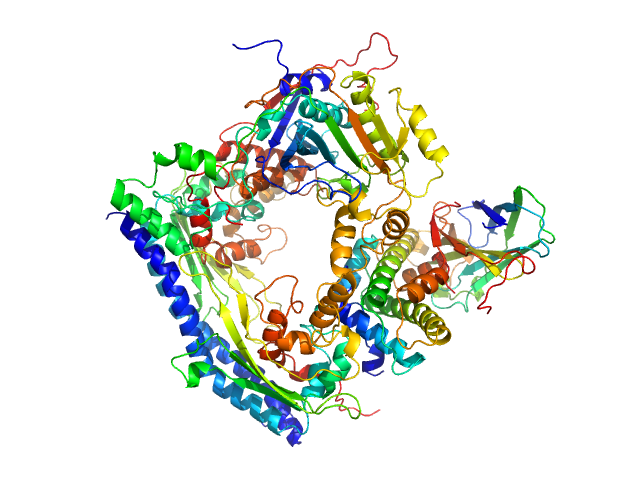
UniProt ID: P53853 (1-225) Vacuolar protein sorting-associated protein 75 (1-225 aa)
UniProt ID: Q07794 (1-436) Histone acetyltransferase RTT109
UniProt ID: P32447 (1-169) Histone chaperone ASF1
UniProt ID: P84233 (35-135) Histone H3.2 (35-135 aa)
UniProt ID: P62799 (1-103) Histone H4
|
|
|
|
| Sample: |
Vacuolar protein sorting-associated protein 75 (1-225 aa) dimer, 53 kDa Saccharomyces cerevisiae protein
Histone acetyltransferase RTT109 monomer, 50 kDa Saccharomyces cerevisiae protein
Histone chaperone ASF1 monomer, 19 kDa protein
Histone H3.2 (35-135 aa) monomer, 12 kDa Xenopus laevis protein
Histone H4 monomer, 11 kDa Xenopus laevis protein
|
| Buffer: |
50 mM citrate, 150 mM NaCl, 5 mM BME, 100% D2O, pH: 6.5 |
| Experiment: |
SANS
data collected at KWS1, FRM2 on 2017 Mar 4
|
Histone chaperone exploits intrinsic disorder to switch acetylation specificity.
Nat Commun 10(1):3435 (2019)
Danilenko N, Lercher L, Kirkpatrick J, Gabel F, Codutti L, Carlomagno T
|
| RgGuinier |
3.3 |
nm |
| Dmax |
10.5 |
nm |
|
|
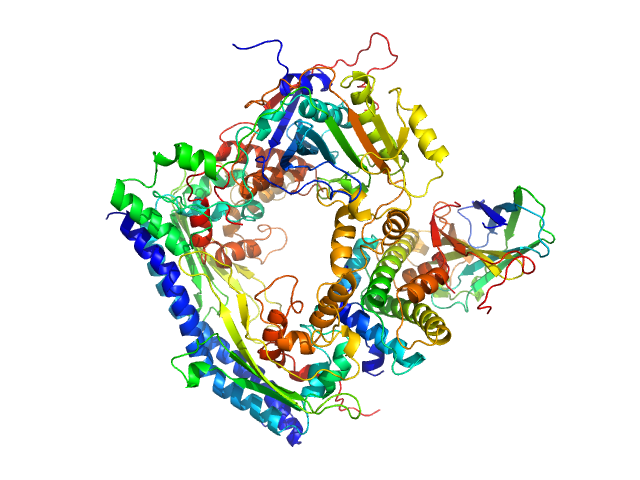
UniProt ID: P53853 (1-225) Vacuolar protein sorting-associated protein 75 (1-225 aa)
UniProt ID: Q07794 (1-436) Histone acetyltransferase RTT109
UniProt ID: P32447 (1-169) Histone chaperone ASF1
UniProt ID: P84233 (35-135) Histone H3.2 (35-135 aa)
UniProt ID: P62799 (1-103) Histone H4
|
|
|
|
| Sample: |
Vacuolar protein sorting-associated protein 75 (1-225 aa) dimer, 53 kDa Saccharomyces cerevisiae protein
Histone acetyltransferase RTT109 monomer, 50 kDa Saccharomyces cerevisiae protein
Histone chaperone ASF1 monomer, 19 kDa protein
Histone H3.2 (35-135 aa) monomer, 12 kDa Xenopus laevis protein
Histone H4 monomer, 11 kDa Xenopus laevis protein
|
| Buffer: |
50 mM citrate, 150 mM NaCl, 5 mM BME, 100% D2O, pH: 6.5 |
| Experiment: |
SANS
data collected at D22, Institut Laue-Langevin (ILL) on 2018 May 29
|
Histone chaperone exploits intrinsic disorder to switch acetylation specificity.
Nat Commun 10(1):3435 (2019)
Danilenko N, Lercher L, Kirkpatrick J, Gabel F, Codutti L, Carlomagno T
|
| RgGuinier |
2.8 |
nm |
| Dmax |
9.5 |
nm |
|
|
UniProt ID: P53853 (1-225) Vacuolar protein sorting-associated protein 75 (1-225 aa)
UniProt ID: Q07794 (1-436) Histone acetyltransferase RTT109
UniProt ID: P32447 (1-169) Histone chaperone ASF1
UniProt ID: P84233 (35-135) Histone H3.2 (35-135 aa)
UniProt ID: P62799 (1-103) Histone H4
|
|
|
|
| Sample: |
Vacuolar protein sorting-associated protein 75 (1-225 aa) dimer, 53 kDa Saccharomyces cerevisiae protein
Histone acetyltransferase RTT109 monomer, 50 kDa Saccharomyces cerevisiae protein
Histone chaperone ASF1 monomer, 19 kDa protein
Histone H3.2 (35-135 aa) monomer, 12 kDa Xenopus laevis protein
Histone H4 monomer, 11 kDa Xenopus laevis protein
|
| Buffer: |
50 mM citrate, 150 mM NaCl, 5 mM BME, 42% D2O, pH: 6.5 |
| Experiment: |
SANS
data collected at KWS1, FRM2 on 2017 Mar 5
|
Histone chaperone exploits intrinsic disorder to switch acetylation specificity.
Nat Commun 10(1):3435 (2019)
Danilenko N, Lercher L, Kirkpatrick J, Gabel F, Codutti L, Carlomagno T
|
| RgGuinier |
3.5 |
nm |
| Dmax |
11.0 |
nm |
|
|
UniProt ID: P53853 (1-225) Vacuolar protein sorting-associated protein 75 (1-225 aa)
UniProt ID: Q07794 (1-436) Histone acetyltransferase RTT109
UniProt ID: P32447 (1-169) Histone chaperone ASF1
UniProt ID: P84233 (35-135) Histone H3.2 (35-135 aa)
UniProt ID: P62799 (1-103) Histone H4
|
|
|
|
| Sample: |
Vacuolar protein sorting-associated protein 75 (1-225 aa) dimer, 53 kDa Saccharomyces cerevisiae protein
Histone acetyltransferase RTT109 monomer, 50 kDa Saccharomyces cerevisiae protein
Histone chaperone ASF1 monomer, 19 kDa protein
Histone H3.2 (35-135 aa) monomer, 12 kDa Xenopus laevis protein
Histone H4 monomer, 11 kDa Xenopus laevis protein
|
| Buffer: |
50 mM citrate, 150 mM NaCl, 5 mM BME, 42% D2O, pH: 6.5 |
| Experiment: |
SANS
data collected at KWS1, FRM2 on 2017 Mar 5
|
Histone chaperone exploits intrinsic disorder to switch acetylation specificity.
Nat Commun 10(1):3435 (2019)
Danilenko N, Lercher L, Kirkpatrick J, Gabel F, Codutti L, Carlomagno T
|
| RgGuinier |
3.1 |
nm |
| Dmax |
10.5 |
nm |
|
|