|
|
|
|
|
| Sample: |
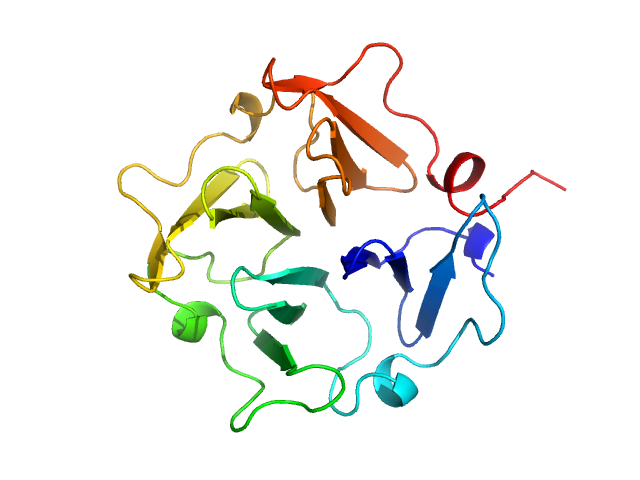
Hemopexin monomer, 23 kDa Homo sapiens protein
Hemopexin dimer, 46 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 150 mM NaCl, 10 mM CaCl2, pH: 7.5
|
| Experiment: |
SAXS
data collected at ...X33, DORIS III, DESY on 2006 Nov 25
|
The Dimer Interface of the Membrane Type 1 Matrix Metalloproteinase Hemopexin Domain
Journal of Biological Chemistry 286(9):7587-7600 (2011)
Tochowicz A, Goettig P, Evans R, Visse R, Shitomi Y, Palmisano R, Ito N, Richter K, Maskos K, Franke D, Svergun D, Nagase H, Bode W, Itoh Y
|
| RgGuinier |
2.3 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
48 |
nm3 |
|