|
|
|
|
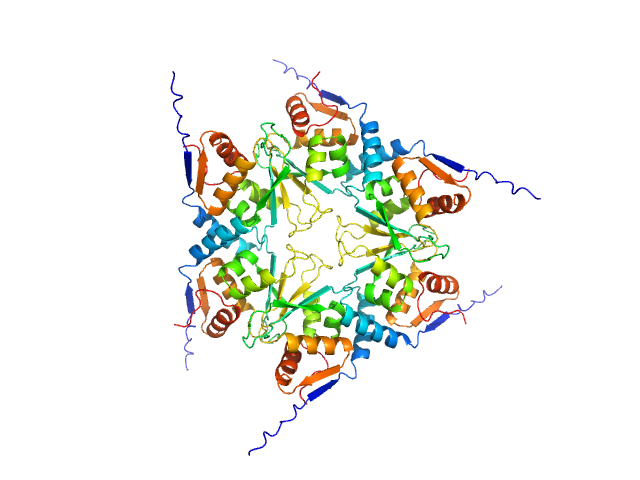
|
| Sample: |
Uncharacterized protein, isoform A hexamer, 92 kDa Drosophila melanogaster protein
|
| Buffer: |
20 mM Tris, pH 7.4, 200 mM NaCl, 1 mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Jul 8
|
BTB domains: A structural view of evolution, multimerization, and protein-protein interactions.
Bioessays :e2200179 (2022)
Bonchuk A, Balagurov K, Georgiev P
|
| RgGuinier |
3.7 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
167 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Uncharacterized protein, isoform A hexamer, 92 kDa Drosophila melanogaster protein
|
| Buffer: |
20 mM Tris, pH 7.4, 200 mM NaCl, 1 mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Jul 8
|
BTB domains: A structural view of evolution, multimerization, and protein-protein interactions.
Bioessays :e2200179 (2022)
Bonchuk A, Balagurov K, Georgiev P
|
| RgGuinier |
4.1 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
192 |
nm3 |
|
|
|
|
|
|
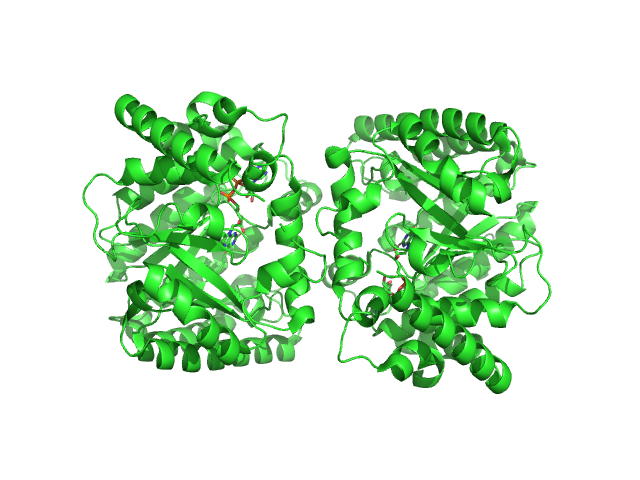
|
| Sample: |
Minimal proline dehydrogenase domain of proline utilization A (design #2) dimer, 87 kDa Sinorhizobium meliloti protein
|
| Buffer: |
25 mM HEPES pH 7.6, 150 mM NaCl, and 1mM TCEP, pH: 7.6 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2022 Apr 12
|
Structure-based engineering of minimal Proline dehydrogenase domains for inhibitor discovery.
Protein Eng Des Sel (2022)
Bogner AN, Ji J, Tanner JJ
|
| RgGuinier |
2.7 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
102 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Minimal proline dehydrogenase domain of proline utilization A (design #2) dimer, 87 kDa Sinorhizobium meliloti protein
|
| Buffer: |
25 mM HEPES pH 7.6, 150 mM NaCl, and 1mM TCEP, pH: 7.6 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2022 Apr 12
|
Structure-based engineering of minimal Proline dehydrogenase domains for inhibitor discovery.
Protein Eng Des Sel (2022)
Bogner AN, Ji J, Tanner JJ
|
| RgGuinier |
2.9 |
nm |
| Dmax |
9.7 |
nm |
| VolumePorod |
102 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Minimal proline dehydrogenase domain of proline utilization A (design #2) dimer, 87 kDa Sinorhizobium meliloti protein
|
| Buffer: |
25 mM HEPES pH 7.6, 150 mM NaCl, and 1mM TCEP, pH: 7.6 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2022 Apr 12
|
Structure-based engineering of minimal Proline dehydrogenase domains for inhibitor discovery.
Protein Eng Des Sel (2022)
Bogner AN, Ji J, Tanner JJ
|
| RgGuinier |
3.0 |
nm |
| Dmax |
9.8 |
nm |
| VolumePorod |
108 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Xylose isomerase tetramer, 173 kDa Streptomyces rubiginosus protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, 3% v/v glycerol, pH: 7 |
| Experiment: |
SAXS
data collected at ID7A1 BioSAXS / HP-Bio Beamline, Cornell High Energy Synchrotron Source (CHESS) on 2020 Mar 7
|
Inline small‐angle X‐ray scattering‐coupled chromatography under extreme hydrostatic pressure
Protein Science 31(12) (2022)
Miller R, Cummings C, Huang Q, Ando N, Gillilan R
|
| RgGuinier |
3.4 |
nm |
| Dmax |
10.6 |
nm |
| VolumePorod |
253 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Xylose isomerase tetramer, 173 kDa Streptomyces rubiginosus protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, and 3% v/v glycerol, pH: 7 |
| Experiment: |
SAXS
data collected at ID7A1 BioSAXS / HP-Bio Beamline, Cornell High Energy Synchrotron Source (CHESS) on 2020 Mar 7
|
Inline small‐angle X‐ray scattering‐coupled chromatography under extreme hydrostatic pressure
Protein Science 31(12) (2022)
Miller R, Cummings C, Huang Q, Ando N, Gillilan R
|
| RgGuinier |
3.5 |
nm |
| Dmax |
10.6 |
nm |
| VolumePorod |
267 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Albumin monomer, 69 kDa Bos taurus protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, and 1% v/v glycerol, pH: 7 |
| Experiment: |
SAXS
data collected at ID7A1 BioSAXS / HP-Bio Beamline, Cornell High Energy Synchrotron Source (CHESS) on 2020 Mar 7
|
Inline small‐angle X‐ray scattering‐coupled chromatography under extreme hydrostatic pressure
Protein Science 31(12) (2022)
Miller R, Cummings C, Huang Q, Ando N, Gillilan R
|
| RgGuinier |
2.7 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
99 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Albumin monomer, 69 kDa Bos taurus protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, and 1% v/v glycerol, pH: 7 |
| Experiment: |
SAXS
data collected at ID7A1 BioSAXS / HP-Bio Beamline, Cornell High Energy Synchrotron Source (CHESS) on 2020 Mar 7
|
Inline small‐angle X‐ray scattering‐coupled chromatography under extreme hydrostatic pressure
Protein Science 31(12) (2022)
Miller R, Cummings C, Huang Q, Ando N, Gillilan R
|
| RgGuinier |
2.6 |
nm |
| Dmax |
8.3 |
nm |
| VolumePorod |
94 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Albumin monomer, 69 kDa Bos taurus protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, and 1% v/v glycerol, pH: 7 |
| Experiment: |
SAXS
data collected at ID7A1 BioSAXS / HP-Bio Beamline, Cornell High Energy Synchrotron Source (CHESS) on 2020 Mar 7
|
Inline small‐angle X‐ray scattering‐coupled chromatography under extreme hydrostatic pressure
Protein Science 31(12) (2022)
Miller R, Cummings C, Huang Q, Ando N, Gillilan R
|
| RgGuinier |
3.0 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
108 |
nm3 |
|
|