|
|
|
|
|
| Sample: |
Albumin monomer, 69 kDa Bos taurus protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, and 1% v/v glycerol, pH: 7 |
| Experiment: |
SAXS
data collected at ID7A1 BioSAXS / HP-Bio Beamline, Cornell High Energy Synchrotron Source (CHESS) on 2020 Mar 7
|
Inline small‐angle X‐ray scattering‐coupled chromatography under extreme hydrostatic pressure
Protein Science 31(12) (2022)
Miller R, Cummings C, Huang Q, Ando N, Gillilan R
|
| RgGuinier |
2.8 |
nm |
| Dmax |
9.4 |
nm |
| VolumePorod |
104 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Acidic leucine-rich nuclear phosphoprotein 32 family member A (L60A mutant) monomer, 29 kDa Homo sapiens protein
|
| Buffer: |
25 mM BisTRIS, 10 mM NaCl, 5 mM DTT, pH: 6.8 |
| Experiment: |
SAXS
data collected at ID7A1 BioSAXS / HP-Bio Beamline, Cornell High Energy Synchrotron Source (CHESS) on 2020 Mar 7
|
Inline small‐angle X‐ray scattering‐coupled chromatography under extreme hydrostatic pressure
Protein Science 31(12) (2022)
Miller R, Cummings C, Huang Q, Ando N, Gillilan R
|
| RgGuinier |
1.8 |
nm |
| Dmax |
6.0 |
nm |
| VolumePorod |
31 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Acidic leucine-rich nuclear phosphoprotein 32 family member A (L60A mutant) monomer, 29 kDa Homo sapiens protein
|
| Buffer: |
25 mM BisTRIS, 10 mM NaCl, 5 mM DTT, pH: 6.8 |
| Experiment: |
SAXS
data collected at ID7A1 BioSAXS / HP-Bio Beamline, Cornell High Energy Synchrotron Source (CHESS) on 2020 Mar 7
|
Inline small‐angle X‐ray scattering‐coupled chromatography under extreme hydrostatic pressure
Protein Science 31(12) (2022)
Miller R, Cummings C, Huang Q, Ando N, Gillilan R
|
| RgGuinier |
2.0 |
nm |
| Dmax |
7.5 |
nm |
| VolumePorod |
37 |
nm3 |
|
|
|
|
|
|
|
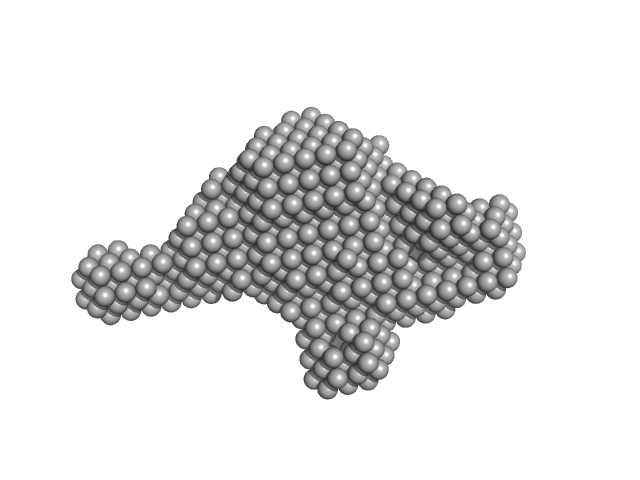
| Sample: |
Ras GTPase-activating protein 1 monomer, 101 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris pH 8 350 mM NaCl 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2020 Dec 12
|
Tandem engagement of phosphotyrosines by the dual SH2 domains of p120RasGAP.
Structure (2022)
Stiegler AL, Vish KJ, Boggon TJ
|
| RgGuinier |
2.6 |
nm |
| Dmax |
10.3 |
nm |
| VolumePorod |
41 |
nm3 |
|
|
|
|
|
|
|
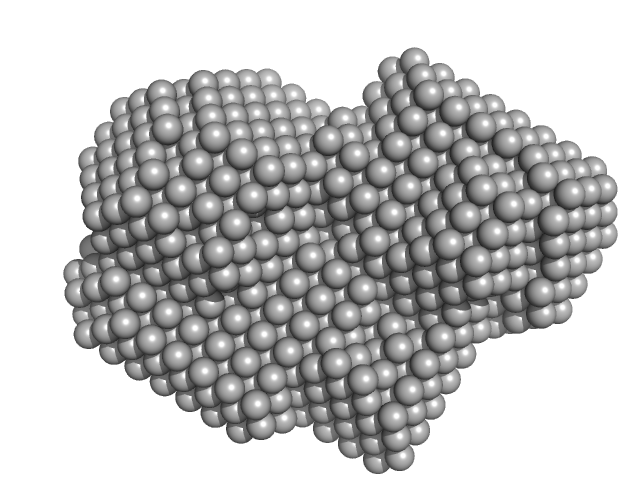
| Sample: |
Ras GTPase-activating protein 1 monomer, 101 kDa Homo sapiens protein
Rho GTPase-activating protein 35 monomer, 3 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris pH 8 350 mM NaCl 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2020 Dec 11
|
Tandem engagement of phosphotyrosines by the dual SH2 domains of p120RasGAP.
Structure (2022)
Stiegler AL, Vish KJ, Boggon TJ
|
| RgGuinier |
2.4 |
nm |
| Dmax |
7.9 |
nm |
| VolumePorod |
41 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
DHPC - 1,2-dihexanoyl-sn-glycero-3-phosphocholine None, lipid
DMPC - 1,2-dimyristoyl-sn-glycero-3-phosphocholine None, lipid
|
| Buffer: |
Tris buffered saline, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Jun 1
|
Expanding the Toolbox for Bicelle-Forming Surfactant–Lipid Mixtures
Molecules 27(21):7628 (2022)
Giudice R, Paracini N, Laursen T, Blanchet C, Roosen-Runge F, Cárdenas M
|
|
|
|
|
|
|
|
| Sample: |
DMPC - 1,2-dimyristoyl-sn-glycero-3-phosphocholine None, lipid
Cholate - 3α,7α,12α-trihydroxy-5β-cholan-24-oic acid None, lipid
|
| Buffer: |
Tris buffered saline, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Mar 17
|
Expanding the Toolbox for Bicelle-Forming Surfactant–Lipid Mixtures
Molecules 27(21):7628 (2022)
Giudice R, Paracini N, Laursen T, Blanchet C, Roosen-Runge F, Cárdenas M
|
|
|
|
|
|
|
|
| Sample: |
DMPC - 1,2-dimyristoyl-sn-glycero-3-phosphocholine None, lipid
CHAPS - 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate None, lipid
|
| Buffer: |
Tris buffered saline, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Mar 17
|
Expanding the Toolbox for Bicelle-Forming Surfactant–Lipid Mixtures
Molecules 27(21):7628 (2022)
Giudice R, Paracini N, Laursen T, Blanchet C, Roosen-Runge F, Cárdenas M
|
|
|
|
|
|
|
|
| Sample: |
DMPC - 1,2-dimyristoyl-sn-glycero-3-phosphocholine None, lipid
CHAPS - 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate None, lipid
DMPG - 1,2-dimyristoyl-sn-glycero-3-phospho-sn-glycerol None, lipid
|
| Buffer: |
Tris buffered saline, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Mar 17
|
Expanding the Toolbox for Bicelle-Forming Surfactant–Lipid Mixtures
Molecules 27(21):7628 (2022)
Giudice R, Paracini N, Laursen T, Blanchet C, Roosen-Runge F, Cárdenas M
|
|
|
|
|
|
|
|
| Sample: |
DMPC - 1,2-dimyristoyl-sn-glycero-3-phosphocholine None, lipid
Cholate - 3α,7α,12α-trihydroxy-5β-cholan-24-oic acid None, lipid
DMPG - 1,2-dimyristoyl-sn-glycero-3-phospho-sn-glycerol None, lipid
|
| Buffer: |
Tris buffered saline, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Mar 17
|
Expanding the Toolbox for Bicelle-Forming Surfactant–Lipid Mixtures
Molecules 27(21):7628 (2022)
Giudice R, Paracini N, Laursen T, Blanchet C, Roosen-Runge F, Cárdenas M
|
|
|