|
|
|
|
|
| Sample: |
NF-kappa-B p105 subunit 39-350 monomer, 36 kDa Mus musculus protein
Transcription factor p65 19-321 monomer, 35 kDa Mus musculus protein
IFN kB DNA monomer, 8 kDa DNA
|
| Buffer: |
25 mM Tris.Cl, 150 mM NaCl, 1 mM DTT, 0.5 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Jun 11
|
An intrinsically disordered transcription activation domain increases the DNA binding affinity and reduces the specificity of NFκB p50/RelA.
J Biol Chem :102349 (2022)
Baughman HER, Narang D, Chen W, Villagrán Suárez AC, Lee J, Bachochin MJ, Gunther TR, Wolynes PG, Komives EA
|
| RgGuinier |
4.3 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
237 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
NF-kappa-B p105 subunit 39-350 monomer, 36 kDa Mus musculus protein
Transcription factor p65 19-321 monomer, 35 kDa Mus musculus protein
Urokinase kB monomer, 8 kDa DNA
|
| Buffer: |
25 mM Tris.Cl, 150 mM NaCl, 1 mM DTT, 0.5 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Jun 11
|
An intrinsically disordered transcription activation domain increases the DNA binding affinity and reduces the specificity of NFκB p50/RelA.
J Biol Chem :102349 (2022)
Baughman HER, Narang D, Chen W, Villagrán Suárez AC, Lee J, Bachochin MJ, Gunther TR, Wolynes PG, Komives EA
|
| RgGuinier |
4.1 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
244 |
nm3 |
|
|
|
|
|
|
|
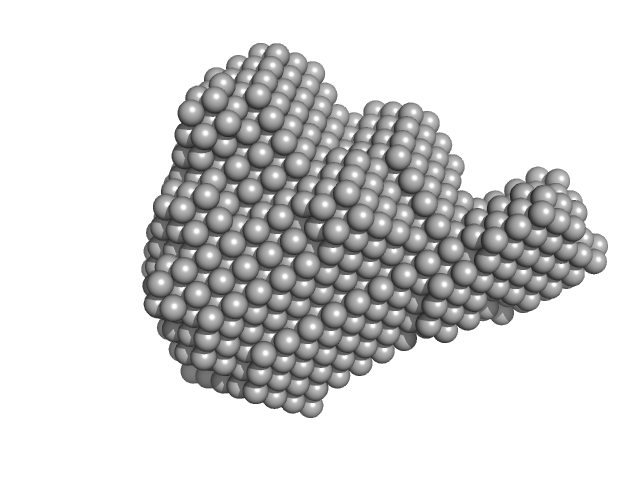
| Sample: |
Transcription factor p65 19-549 monomer, 58 kDa Mus musculus protein
NF-kappa-B p105 subunit 39-350 monomer, 36 kDa Mus musculus protein
IFN kB DNA monomer, 8 kDa DNA
|
| Buffer: |
25 mM Tris.Cl, 150 mM NaCl, 1 mM DTT, 0.5 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Dec 4
|
An intrinsically disordered transcription activation domain increases the DNA binding affinity and reduces the specificity of NFκB p50/RelA.
J Biol Chem :102349 (2022)
Baughman HER, Narang D, Chen W, Villagrán Suárez AC, Lee J, Bachochin MJ, Gunther TR, Wolynes PG, Komives EA
|
| RgGuinier |
5.1 |
nm |
| Dmax |
18.9 |
nm |
| VolumePorod |
314 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Transcription factor p65 19-549 monomer, 58 kDa Mus musculus protein
NF-kappa-B p105 subunit 39-350 monomer, 36 kDa Mus musculus protein
Urokinase kB monomer, 8 kDa DNA
|
| Buffer: |
25 mM Tris.Cl, 150 mM NaCl, 1 mM DTT, 0.5 mM EDTA, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Dec 4
|
An intrinsically disordered transcription activation domain increases the DNA binding affinity and reduces the specificity of NFκB p50/RelA.
J Biol Chem :102349 (2022)
Baughman HER, Narang D, Chen W, Villagrán Suárez AC, Lee J, Bachochin MJ, Gunther TR, Wolynes PG, Komives EA
|
| RgGuinier |
4.7 |
nm |
| Dmax |
17.5 |
nm |
| VolumePorod |
201 |
nm3 |
|
|
|
|
|
|
|
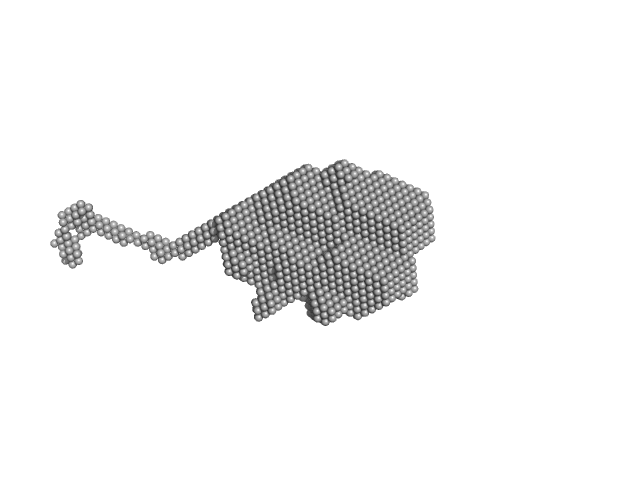
| Sample: |
Aprataxin and PNK-like factor (acidic domain) dimer, 15 kDa Homo sapiens protein
Histone H2A dimer, 26 kDa Drosophila melanogaster protein
Histone H2B dimer, 27 kDa Drosophila melanogaster protein
Histone H3 dimer, 30 kDa Drosophila melanogaster protein
Histone H4 dimer, 23 kDa Drosophila melanogaster protein
|
| Buffer: |
25 mM NaPi, 300 mM NaCl, 3% v/v glycerol, 1 mM DTT,, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Jul 10
|
Chaperoning of the histone octamer by the acidic domain of DNA repair factor APLF.
Sci Adv 8(30):eabo0517 (2022)
Corbeski I, Guo X, Eckhardt BV, Fasci D, Wiegant W, Graewert MA, Vreeken K, Wienk H, Svergun DI, Heck AJR, van Attikum H, Boelens R, Sixma TK, Mattiroli F, van Ingen H
|
| RgGuinier |
3.7 |
nm |
| Dmax |
10.8 |
nm |
| VolumePorod |
260 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Protein DPCD monomer, 24 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 1% glycerol, 5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Mar 21
|
Deciphering cellular and molecular determinants of human DPCD protein in complex with RUVBL1/RUVBL2 AAA-ATPases.
J Mol Biol :167760 (2022)
Dos Santos Morais R, Santo PE, Ley M, Schelcher C, Abel Y, Plassart L, Deslignière E, Chagot ME, Quinternet M, Paiva ACF, Hessmann S, Morellet N, M F Sousa P, Vandermoere F, Bertrand E, Charpentier B, Bandeiras TM, Plisson-Chastang C, Verheggen C, Cianférani S, Manival X
|
| RgGuinier |
2.2 |
nm |
| Dmax |
12.3 |
nm |
| VolumePorod |
39 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
RuvB-like 1 hexamer, 311 kDa Homo sapiens protein
RuvB-like 2 hexamer, 311 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 1% glycerol, 5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Mar 21
|
Deciphering cellular and molecular determinants of human DPCD protein in complex with RUVBL1/RUVBL2 AAA-ATPases.
J Mol Biol :167760 (2022)
Dos Santos Morais R, Santo PE, Ley M, Schelcher C, Abel Y, Plassart L, Deslignière E, Chagot ME, Quinternet M, Paiva ACF, Hessmann S, Morellet N, M F Sousa P, Vandermoere F, Bertrand E, Charpentier B, Bandeiras TM, Plisson-Chastang C, Verheggen C, Cianférani S, Manival X
|
| RgGuinier |
6.2 |
nm |
| Dmax |
20.0 |
nm |
| VolumePorod |
1490 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Protein DPCD trimer, 71 kDa Homo sapiens protein
RuvB-like 1 trimer, 155 kDa Homo sapiens protein
RuvB-like 2 trimer, 156 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 1% glycerol, 5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Mar 21
|
Deciphering cellular and molecular determinants of human DPCD protein in complex with RUVBL1/RUVBL2 AAA-ATPases.
J Mol Biol :167760 (2022)
Dos Santos Morais R, Santo PE, Ley M, Schelcher C, Abel Y, Plassart L, Deslignière E, Chagot ME, Quinternet M, Paiva ACF, Hessmann S, Morellet N, M F Sousa P, Vandermoere F, Bertrand E, Charpentier B, Bandeiras TM, Plisson-Chastang C, Verheggen C, Cianférani S, Manival X
|
| RgGuinier |
5.4 |
nm |
| Dmax |
15.9 |
nm |
| VolumePorod |
856 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Lysin [Streptococcus phage P7951] hexamer, 65 kDa Streptococcus phage P7951 protein
|
| Buffer: |
50 mM HEPES, 500 mM NaCl, and 1% glycerol, pH: 7 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2021 Nov 21
|
On the Occurrence and Multimerization of Two-Polypeptide Phage Endolysins Encoded in Single Genes.
Microbiol Spectr :e0103722 (2022)
Pinto D, Gonçalo R, Louro M, Silva MS, Hernandez G, Cordeiro TN, Cordeiro C, São-José C
|
| RgGuinier |
3.1 |
nm |
| Dmax |
10.1 |
nm |
| VolumePorod |
111 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Lipid II isoglutaminyl synthase (glutamine-hydrolyzing) subunit MurT monomer, 52 kDa Streptococcus pneumoniae (strain … protein
Lipid II isoglutaminyl synthase (glutamine-hydrolyzing) subunit GatD monomer, 29 kDa Streptococcus pneumoniae (strain … protein
|
| Buffer: |
50 mM Hepes, 10 mM MgCl2, 500 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Jun 23
|
Unravelling the reaction mechanism of glutamate amidation in Staphylococcus aureus peptidoglycan
PhD thesis, NOVA University Lisbon - (2022)
Francisco Miguel Piçarra Leisico, Mertens HD
|
| RgGuinier |
3.0 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
126 |
nm3 |
|
|