|
|
|
|

|
| Sample: |
Alpha/beta fold hydrolase monomer, 28 kDa Staphylococcus aureus protein
|
| Buffer: |
100 mM NaCl, 10 mM HEPES, pH: 7.6 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2024 Apr 11
|
Similar but Distinct—Biochemical Characterization of the
Staphylococcus aureus
Serine Hydrolases FphH
and FphI
Proteins: Structure, Function, and Bioinformatics (2024)
Fellner M, Randall G, Bitac I, Warrender A, Sethi A, Jelinek R, Kass I
|
| RgGuinier |
2.1 |
nm |
| Dmax |
6.5 |
nm |
| VolumePorod |
41 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Partner and localizer of BRCA2 dimer, 46 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 500 mM NaCl, 0.5 mM TCEP, pH: 7.4 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Nov 11
|
The strand exchange domain of tumor suppressor PALB2 is intrinsically disordered and promotes oligomerization-dependent DNA compaction.
iScience 27(12):111259 (2024)
Kyriukha Y, Watkins MB, Redington JM, Chintalapati N, Ganti A, Dastvan R, Uversky VN, Hopkins JB, Pozzi N, Korolev S
|
| RgGuinier |
5.5 |
nm |
| Dmax |
25.4 |
nm |
| VolumePorod |
171 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Partner and localizer of BRCA2 dimer, 46 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 160 mM NaCl, 0.5 mM TCEP, pH: 7.4 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Nov 11
|
The strand exchange domain of tumor suppressor PALB2 is intrinsically disordered and promotes oligomerization-dependent DNA compaction.
iScience 27(12):111259 (2024)
Kyriukha Y, Watkins MB, Redington JM, Chintalapati N, Ganti A, Dastvan R, Uversky VN, Hopkins JB, Pozzi N, Korolev S
|
| RgGuinier |
4.7 |
nm |
| Dmax |
21.3 |
nm |
| VolumePorod |
197 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Partner and localizer of BRCA2 (L24A) monomer, 23 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 160 mM NaCl, 0.5 mM TCEP pH 7.5, pH: 7.5 |
| Experiment: |
SAXS
data collected at ID7A1 BioSAXS / HP-Bio Beamline, Cornell High Energy Synchrotron Source (CHESS) on 2023 Dec 9
|
The strand exchange domain of tumor suppressor PALB2 is intrinsically disordered and promotes oligomerization-dependent DNA compaction.
iScience 27(12):111259 (2024)
Kyriukha Y, Watkins MB, Redington JM, Chintalapati N, Ganti A, Dastvan R, Uversky VN, Hopkins JB, Pozzi N, Korolev S
|
| RgGuinier |
5.2 |
nm |
| Dmax |
21.6 |
nm |
| VolumePorod |
127 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Partner and localizer of BRCA2 dimer, 46 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 160 mM NaCl, 0.5 mM TCEP pH 7.5, pH: 7.5 |
| Experiment: |
SAXS
data collected at ID7A1 BioSAXS / HP-Bio Beamline, Cornell High Energy Synchrotron Source (CHESS) on 2023 Dec 9
|
The strand exchange domain of tumor suppressor PALB2 is intrinsically disordered and promotes oligomerization-dependent DNA compaction.
iScience 27(12):111259 (2024)
Kyriukha Y, Watkins MB, Redington JM, Chintalapati N, Ganti A, Dastvan R, Uversky VN, Hopkins JB, Pozzi N, Korolev S
|
| RgGuinier |
4.6 |
nm |
| Dmax |
17.6 |
nm |
| VolumePorod |
236 |
nm3 |
|
|
|
|
|
|
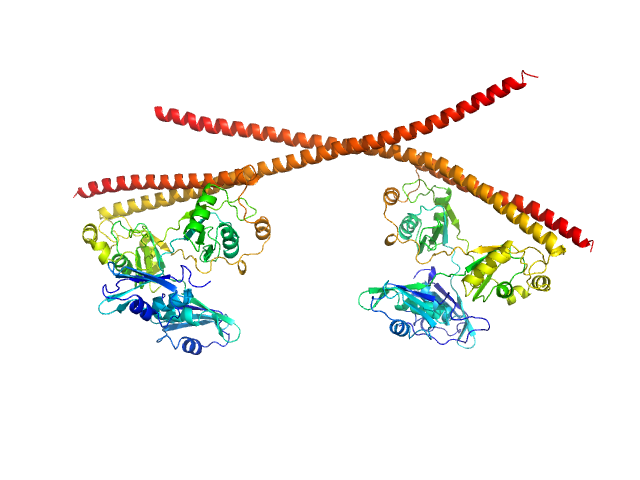
|
| Sample: |
Splicing factor, proline- and glutamine-rich dimer, 153 kDa Homo sapiens protein
Non-POU domain-containing octamer-binding protein dimer, 60 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris pH 7.5, 250 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2019 Jun 18
|
Structural plasticity of the coiled-coil interactions in human SFPQ.
Nucleic Acids Res (2024)
Koning HJ, Lai JY, Marshall AC, Stroeher E, Monahan G, Pullakhandam A, Knott GJ, Ryan TM, Fox AH, Whitten A, Lee M, Bond CS
|
| RgGuinier |
5.5 |
nm |
| Dmax |
20.4 |
nm |
| VolumePorod |
304 |
nm3 |
|
|
|
|
|
|
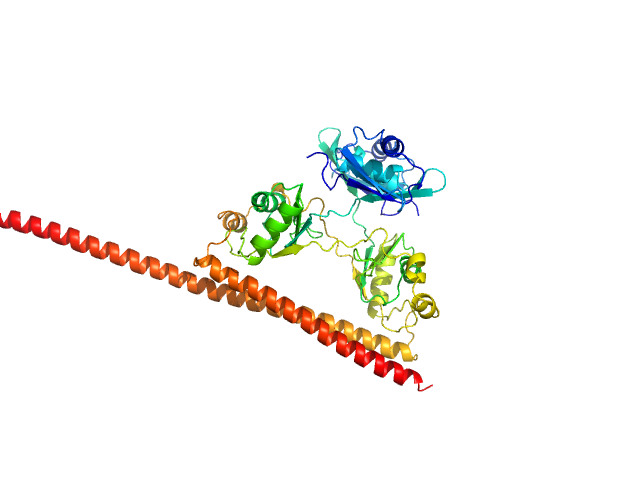
|
| Sample: |
Splicing factor, proline- and glutamine-rich (276-565) monomer, 34 kDa Homo sapiens protein
Non-POU domain-containing octamer-binding protein (53-312) monomer, 30 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris pH 7.5, 250 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2015 Apr 28
|
Structural plasticity of the coiled-coil interactions in human SFPQ.
Nucleic Acids Res (2024)
Koning HJ, Lai JY, Marshall AC, Stroeher E, Monahan G, Pullakhandam A, Knott GJ, Ryan TM, Fox AH, Whitten A, Lee M, Bond CS
|
| RgGuinier |
3.0 |
nm |
| Dmax |
12.7 |
nm |
| VolumePorod |
102 |
nm3 |
|
|
|
|
|
|
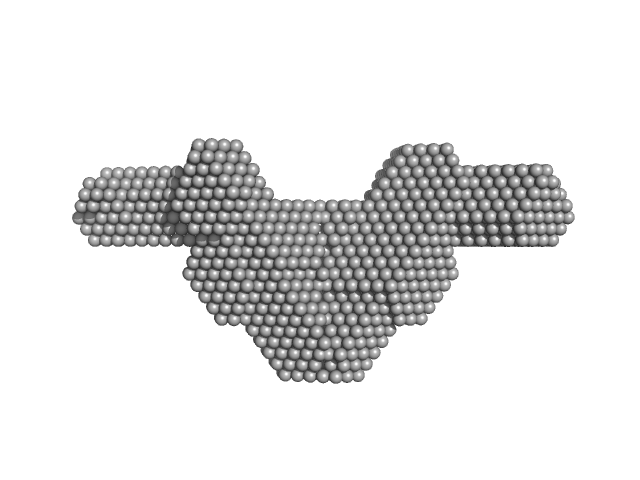
|
| Sample: |
Non-POU domain-containing octamer-binding protein dimer, 60 kDa Homo sapiens protein
Splicing factor, proline- and glutamine-rich SFPQ276-598(R542C)/NONO53-312 dimer) dimer, 76 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris pH 7.5, 250 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2015 Apr 28
|
Structural plasticity of the coiled-coil interactions in human SFPQ.
Nucleic Acids Res (2024)
Koning HJ, Lai JY, Marshall AC, Stroeher E, Monahan G, Pullakhandam A, Knott GJ, Ryan TM, Fox AH, Whitten A, Lee M, Bond CS
|
| RgGuinier |
5.2 |
nm |
| Dmax |
21.0 |
nm |
| VolumePorod |
257 |
nm3 |
|
|
|
|
|
|
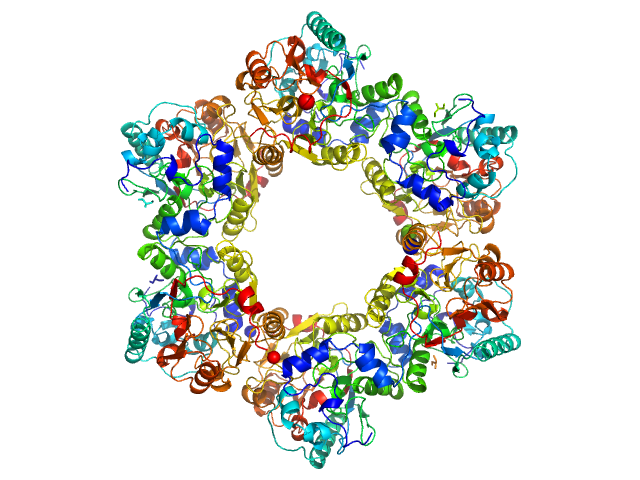
|
| Sample: |
2-nitroimidazole nitrohydrolase (T2I, G14D, K73R) hexamer, 258 kDa Mycobacterium sp. (strain … protein
|
| Buffer: |
25 mM Tris-HCl, 150 mM NaCl, 5% (v/v) glycerol, pH: 7.6 |
| Experiment: |
SAXS
data collected at BioSAXS, Australian Synchrotron on 2024 Aug 6
|
Structural insights into the enzymatic breakdown of azomycin-derived antibiotics by 2-nitroimdazole hydrolase (NnhA).
Commun Biol 7(1):1676 (2024)
Ahmed FH, Liu JW, Royan S, Warden AC, Esquirol L, Pandey G, Newman J, Scott C, Peat TS
|
| RgGuinier |
4.6 |
nm |
| Dmax |
12.6 |
nm |
| VolumePorod |
363 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
RNase P RNA monomer, 135 kDa Geobacillus stearothermophilus RNA
|
| Buffer: |
100 mM NaCl, 0.1 mM MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at Rigaku BIOSAXS-2000 [duplicate], Center for Cancer Research,National Cancer Institute on 2023 Nov 3
|
The conformational space of RNase P RNA in solution
Nature (2024)
Lee Y, Degenhardt M, Skeparnias I, Degenhardt H, Bhandari Y, Yu P, Stagno J, Fan L, Zhang J, Wang Y
|
| RgGuinier |
6.1 |
nm |
| Dmax |
21.8 |
nm |
| VolumePorod |
813 |
nm3 |
|
|