|
|
|
|
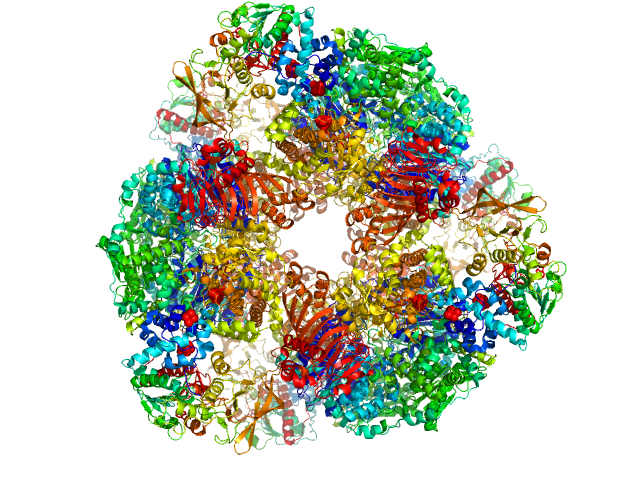
|
| Sample: |
Glutamate synthase [NADPH] large chain hexamer, 968 kDa Azospirillum brasilense protein
Glutamate synthase [NADPH] small chain hexamer, 296 kDa Azospirillum brasilense protein
|
| Buffer: |
25 mM Hepes/KOH, 1 mM EDTA, 1 mM dithiothreitol, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2007 Feb 19
|
The Subnanometer Resolution Structure of the Glutamate Synthase 1.2-MDa Hexamer by Cryoelectron Microscopy and Its Oligomerization Behavior in Solution
Journal of Biological Chemistry 283(13):8237-8249 (2008)
Cottevieille M, Larquet E, Jonic S, Petoukhov M, Caprini G, Paravisi S, Svergun D, Vanoni M, Boisset N
|
| RgGuinier |
7.7 |
nm |
| Dmax |
22.4 |
nm |
|
|
|
|
|
|
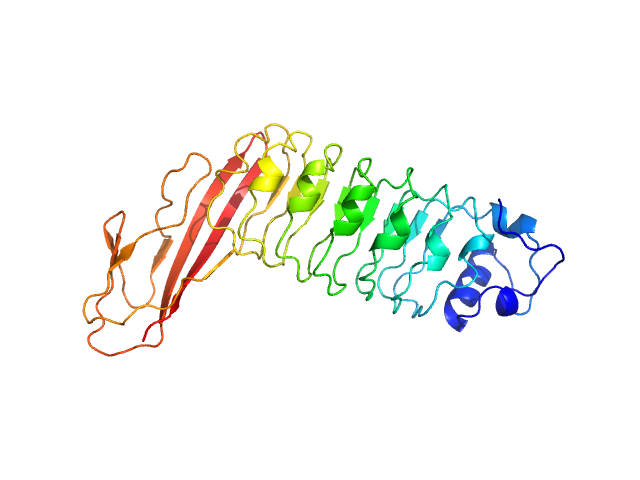
|
| Sample: |
Internalin B monomer, 36 kDa Listeria monocytogenes serotype … protein
|
| Buffer: |
25mM Na-phosphate buffer, 150 mM NaCl, 1mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2003 Nov 11
|
X-ray and Neutron Small-Angle Scattering Analysis of the Complex Formed by the Met Receptor and the Listeria monocytogenes Invasion Protein InlB
Journal of Molecular Biology 377(2):489-500 (2008)
Niemann H, Petoukhov M, Härtlein M, Moulin M, Gherardi E, Timmins P, Heinz D, Svergun D
|
| RgGuinier |
2.8 |
nm |
| Dmax |
9.6 |
nm |
| VolumePorod |
54 |
nm3 |
|
|
|
|
|
|
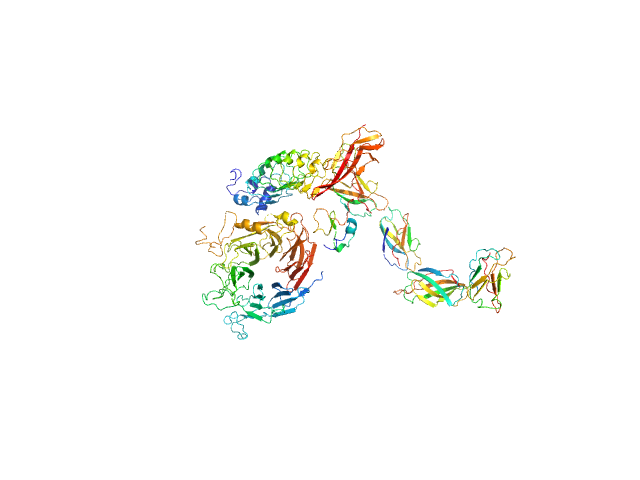
|
| Sample: |
Hepatocyte growth factor receptor monomer, 100 kDa Mus musculus protein
Internalin B monomer, 36 kDa Listeria monocytogenes serotype … protein
|
| Buffer: |
25mM Na-phosphate buffer, 150 mM NaCl, 1mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2004 Mar 1
|
X-ray and Neutron Small-Angle Scattering Analysis of the Complex Formed by the Met Receptor and the Listeria monocytogenes Invasion Protein InlB
Journal of Molecular Biology 377(2):489-500 (2008)
Niemann H, Petoukhov M, Härtlein M, Moulin M, Gherardi E, Timmins P, Heinz D, Svergun D
|
| RgGuinier |
4.6 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
270 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Fab fragment in complex with small molecule hapten, crystal form-1 monomer, 45 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris–HCl, 50 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2005 Jul 25
|
Fab MOR03268 Triggers Absorption Shift of a Diagnostic Dye via Packaging in a Solvent-shielded Fab Dimer Interface
Journal of Molecular Biology 377(1):206-219 (2008)
Hillig R, Urlinger S, Fanghänel J, Brocks B, Haenel C, Stark Y, Sülzle D, Svergun D, Baesler S, Malawski G, Moosmayer D, Menrad A, Schirner M, Licha K
|
| RgGuinier |
3.0 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
71 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Fab fragment in complex with small molecule hapten, crystal form-1 monomer, 45 kDa Homo sapiens protein
(1S)-1-AMINO-2-(1H-INDOL-3-YL)ETHANOL monomer, 0 kDa
|
| Buffer: |
20 mM Tris–HCl, 50 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2005 Jul 27
|
Fab MOR03268 Triggers Absorption Shift of a Diagnostic Dye via Packaging in a Solvent-shielded Fab Dimer Interface
Journal of Molecular Biology 377(1):206-219 (2008)
Hillig R, Urlinger S, Fanghänel J, Brocks B, Haenel C, Stark Y, Sülzle D, Svergun D, Baesler S, Malawski G, Moosmayer D, Menrad A, Schirner M, Licha K
|
| RgGuinier |
4.1 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
139 |
nm3 |
|
|
|
|
|
|
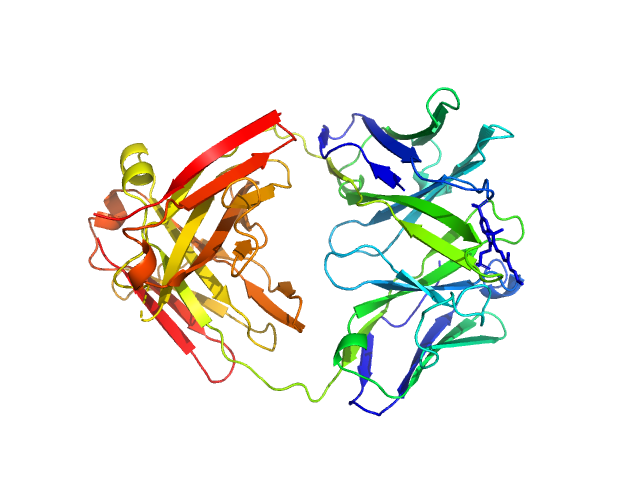
|
| Sample: |
Fab fragment in complex with small molecule hapten, crystal form-1 monomer, 45 kDa Homo sapiens protein
(1S)-1-AMINO-2-(1H-INDOL-3-YL)ETHANOL monomer, 0 kDa
|
| Buffer: |
20 mM Tris–HCl, 50 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2005 Jul 25
|
Fab MOR03268 Triggers Absorption Shift of a Diagnostic Dye via Packaging in a Solvent-shielded Fab Dimer Interface
Journal of Molecular Biology 377(1):206-219 (2008)
Hillig R, Urlinger S, Fanghänel J, Brocks B, Haenel C, Stark Y, Sülzle D, Svergun D, Baesler S, Malawski G, Moosmayer D, Menrad A, Schirner M, Licha K
|
| RgGuinier |
3.6 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
107 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Calmodulin monomer, 17 kDa Homo sapiens protein
C-terminal region of human myelin basic protein monomer, 2 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris75 200 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2006 Nov 28
|
Interaction between the C-terminal region of human myelin basic protein and calmodulin: analysis of complex formation and solution structure.
BMC Struct Biol 8:10 (2008)
Majava V, Petoukhov MV, Hayashi N, Pirilä P, Svergun DI, Kursula P
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.0 |
nm |
| VolumePorod |
36 |
nm3 |
|
|
|
|
|
|
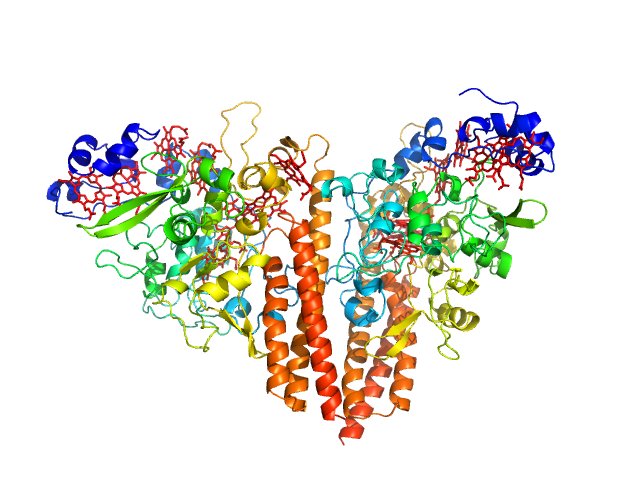
|
| Sample: |
Cytochrome c-552 hexamer, 356 kDa Thioalkalivibrio nitratireducens (strain … protein
|
| Buffer: |
Tris-borate buffer, pH: 8.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2005 May 8
|
Isolation and oligomeric composition of cytochrome c nitrite reductase from the haloalkaliphilic bacterium Thioalkalivibrio nitratireducens.
Biochemistry (Mosc) 73(2):164-70 (2008)
Tikhonova TV, Slutskaya ES, Filimonenkov AA, Boyko KM, Kleimenov SY, Konarev PV, Polyakov KM, Svergun DI, Trofimov AA, Khomenkov VG, Zvyagilskaya RA, Popov VO
|
|
|
|
|
|
|
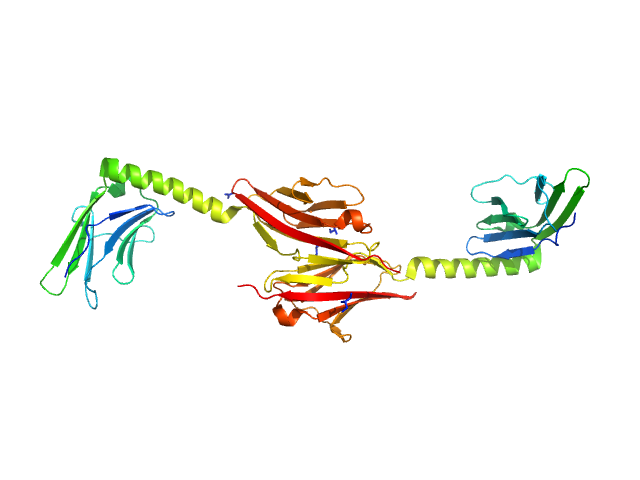
|
| Sample: |
Myomesin-1 dimer, 46 kDa Homo sapiens protein
|
| Buffer: |
25 mM Tris/HCl 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2005 May 7
|
Molecular basis of the C-terminal tail-to-tail assembly of the sarcomeric filament protein myomesin.
EMBO J 27(1):253-64 (2008)
Pinotsis N, Lange S, Perriard JC, Svergun DI, Wilmanns M
|
| RgGuinier |
4.0 |
nm |
| Dmax |
14.5 |
nm |
| VolumePorod |
56 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Human hemoglobin conjugated with six-seven copies of 5-kDa PEG dimer, 62 kDa Homo sapiens protein
|
| Buffer: |
Ringer's lactate solution, pH: 6.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2006 Feb 19
|
Solution Structure of Poly(ethylene) Glycol-Conjugated Hemoglobin Revealed by Small-Angle X-Ray Scattering: Implications for a New Oxygen Therapeutic
Biophysical Journal 94(1):173-181 (2008)
Svergun D, Ekström F, Vandegriff K, Malavalli A, Baker D, Nilsson C, Winslow R
|
|
|