|
|
|
|

|
| Sample: |
Receptor protein-tyrosine kinase (duplication mutant) monomer, 80 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 250 mM NaCl, 250 mM KCl, pH: 8 |
| Experiment: |
SAXS
data collected at Rigaku MicroMax-007HF, Yale University on 2022 Mar 22
|
The role of kinase domain dimerization in EGFR activation
Zaritza Petrova
|
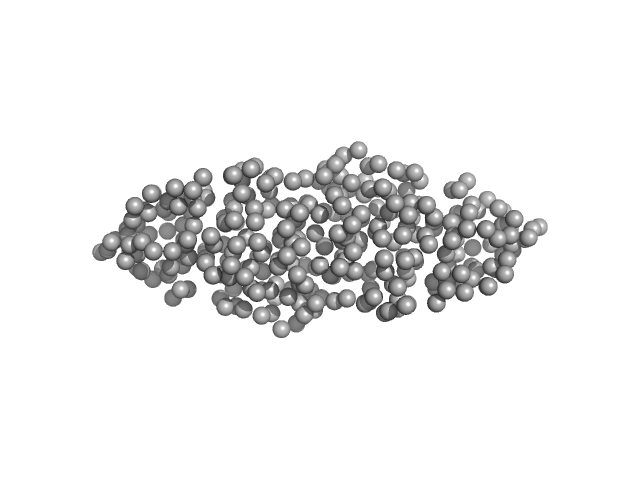
| RgGuinier |
3.9 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
135 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Mutual gliding motility protein C dimer, 32 kDa Myxococcus xanthus (strain … protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 10% glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2023 Mar 31
|
Structural and biophysical insights into RomR, MglB and MglC interactions involved in regulating cell polarity in Myxococcus xanthus
Journal of Biological Chemistry :110907 (2025)
Kodesia A, Kapoor S, Thakur K
|
| RgGuinier |
2.3 |
nm |
| Dmax |
9.8 |
nm |
| VolumePorod |
49 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Two-component system response regulator trimer, 26 kDa Myxococcus xanthus DK … protein
Mutual gliding motility protein C dimer, 32 kDa Myxococcus xanthus (strain … protein
Gliding motility protein MglB dimer, 32 kDa Myxococcus xanthus (strain … protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 10% glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2023 Mar 31
|
Structural and biophysical insights into RomR, MglB and MglC interactions involved in regulating cell polarity in Myxococcus xanthus
Journal of Biological Chemistry :110907 (2025)
Kodesia A, Kapoor S, Thakur K
|
| RgGuinier |
3.8 |
nm |
| Dmax |
13.6 |
nm |
| VolumePorod |
204 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
AH receptor-interacting protein (R9Q) monomer, 39 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris-Cl pH 7.5, 100 mM NaCl, 1 mM DTT, 5% (v/v) glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2018 Jun 13
|
Human AIP HSP90 complex
Robert Rambo
|
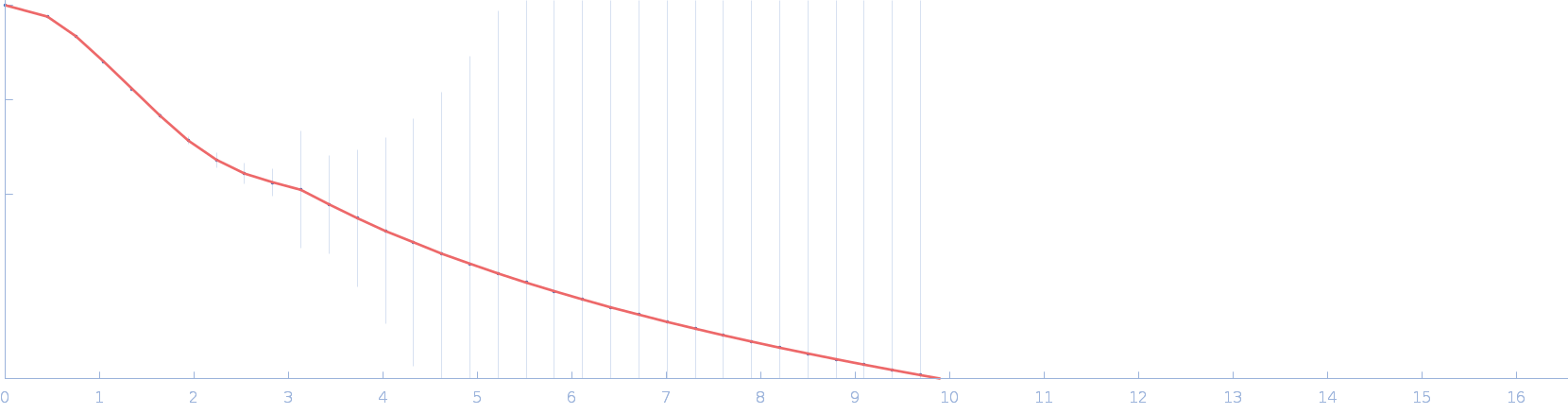
| RgGuinier |
3.0 |
nm |
| Dmax |
10.9 |
nm |
| VolumePorod |
64 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Cellular communication network factor 2 monomer, 36 kDa protein
|
| Buffer: |
50 mM Tris pH 7.5, 200 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 May 6
|
Structural basis for therapeutic antibody binding to CCN2 and its mechanisms on TGF-β family signaling
Haben Gabir
|
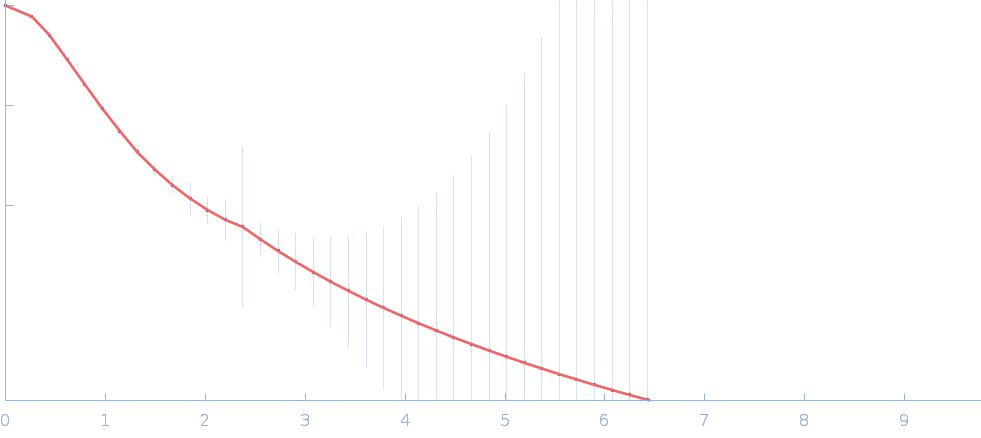
| RgGuinier |
2.1 |
nm |
| Dmax |
7.1 |
nm |
| VolumePorod |
29 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Cellular communication network factor 2 monomer, 36 kDa protein
Anti-CCN2 37-45-MH1 F(ab) antibody monomer, 49 kDa protein
|
| Buffer: |
50 mM Tris pH 7.5, 200 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 May 6
|
Structural basis for therapeutic antibody binding to CCN2 and its mechanisms on TGF-β family signaling
Haben Gabir
|
| RgGuinier |
3.3 |
nm |
| Dmax |
11.9 |
nm |
| VolumePorod |
95 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
UvrABC system protein A dimer, 212 kDa Mycobacterium tuberculosis (strain … protein
UvrABC system protein B dimer, 161 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
50 mM Tris pH 8, 200 mM NaCl, 2% glycerol, pH: |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR-Central Drug Research Institute on 2022 Jun 6
|
The UvrA2B2 pre-initiation complex from the Nucleotide Excision Repair pathway of Mycobacterium tuberculosis.
farheen jahan
|
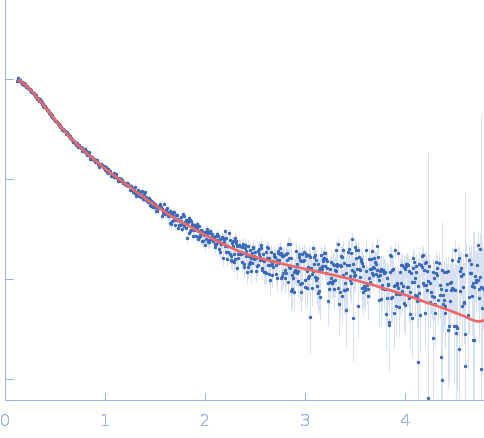
| RgGuinier |
5.3 |
nm |
| Dmax |
15.8 |
nm |
| VolumePorod |
1053 |
nm3 |
|
|
|
|
|
|
|
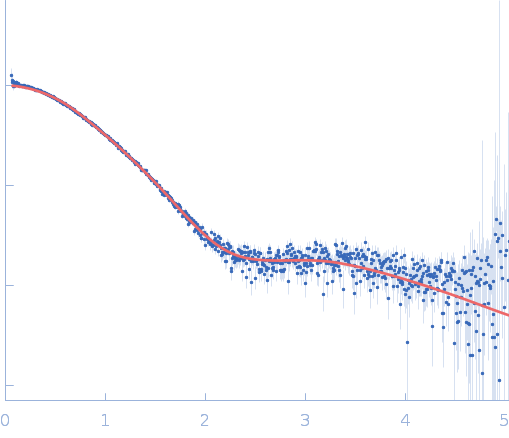
| Sample: |
Repeats-in-toxin domain Block V of adenylate cyclase toxin monomer, 19 kDa Bordetella pertussis protein
|
| Buffer: |
50 mM Tris, 5 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2024 Apr 1
|
Ion-selective conformational stabilization of a disordered repeats-in-toxin protein domain.
Biophys J (2025)
Gudinas AP, Shambharkar GM, Chang MP, Fernández D, Matsui T, Mai DJ
|
| RgGuinier |
3.7 |
nm |
| Dmax |
15.4 |
nm |
| VolumePorod |
52 |
nm3 |
|
|
|
|
|
|
|
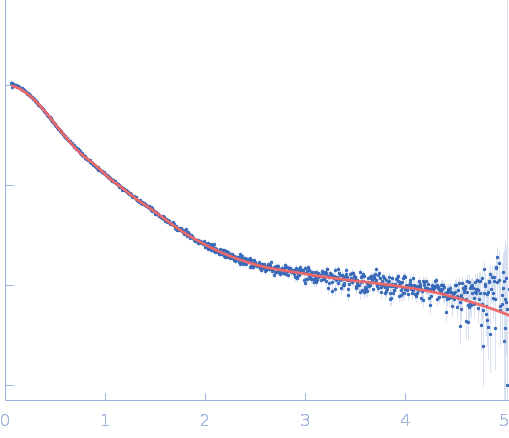
| Sample: |
Repeats-in-toxin domain Block V of adenylate cyclase toxin monomer, 19 kDa Bordetella pertussis protein
|
| Buffer: |
50 mM Tris, 5 mM DTT, 1 mM CaCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2024 Jun 28
|
Ion-selective conformational stabilization of a disordered repeats-in-toxin protein domain.
Biophys J (2025)
Gudinas AP, Shambharkar GM, Chang MP, Fernández D, Matsui T, Mai DJ
|
| RgGuinier |
2.0 |
nm |
| Dmax |
8.1 |
nm |
| VolumePorod |
38 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Repeats-in-toxin domain Block V of adenylate cyclase toxin monomer, 19 kDa Bordetella pertussis protein
|
| Buffer: |
50 mM Tris, 5 mM DTT, 1 mM MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2024 Jun 28
|
Ion-selective conformational stabilization of a disordered repeats-in-toxin protein domain.
Biophys J (2025)
Gudinas AP, Shambharkar GM, Chang MP, Fernández D, Matsui T, Mai DJ
|
| RgGuinier |
3.5 |
nm |
| Dmax |
14.9 |
nm |
| VolumePorod |
49 |
nm3 |
|
|