|
|
|
|
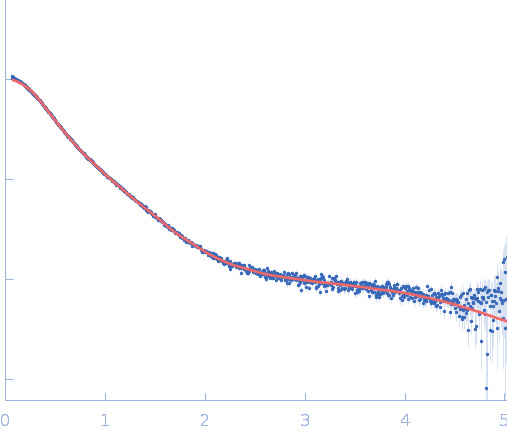
|
| Sample: |
Repeats-in-toxin domain Block V of adenylate cyclase toxin monomer, 19 kDa Bordetella pertussis protein
|
| Buffer: |
50 mM Tris, 5 mM DTT, 10 mM MgCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2024 Aug 1
|
Ion-selective conformational stabilization of a disordered repeats-in-toxin protein domain.
Biophys J (2025)
Gudinas AP, Shambharkar GM, Chang MP, Fernández D, Matsui T, Mai DJ
|
| RgGuinier |
3.8 |
nm |
| Dmax |
16.3 |
nm |
| VolumePorod |
61 |
nm3 |
|
|
|
|
|
|
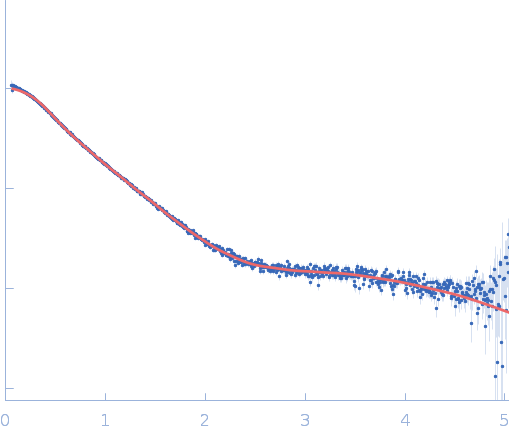
|
| Sample: |
Repeats-in-toxin domain Block V of adenylate cyclase toxin monomer, 19 kDa Bordetella pertussis protein
|
| Buffer: |
50 mM Tris, 5 mM DTT, 1 mM SrCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2024 Jun 28
|
Ion-selective conformational stabilization of a disordered repeats-in-toxin protein domain.
Biophys J (2025)
Gudinas AP, Shambharkar GM, Chang MP, Fernández D, Matsui T, Mai DJ
|
| RgGuinier |
3.1 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
35 |
nm3 |
|
|
|
|
|
|
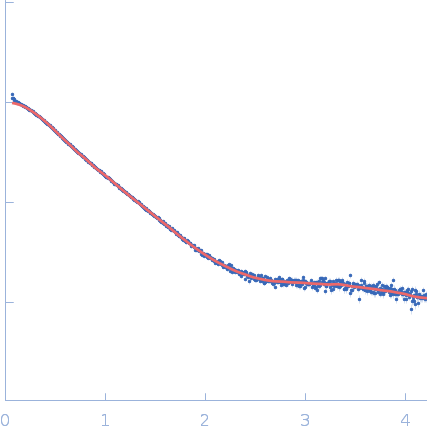
|
| Sample: |
Repeats-in-toxin domain Block V of adenylate cyclase toxin monomer, 19 kDa Bordetella pertussis protein
|
| Buffer: |
50 mM Tris, 5 mM DTT, 1 mM BaCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2024 Aug 1
|
Ion-selective conformational stabilization of a disordered repeats-in-toxin protein domain.
Biophys J (2025)
Gudinas AP, Shambharkar GM, Chang MP, Fernández D, Matsui T, Mai DJ
|
| RgGuinier |
3.0 |
nm |
| Dmax |
13.1 |
nm |
| VolumePorod |
32 |
nm3 |
|
|
|
|
|
|
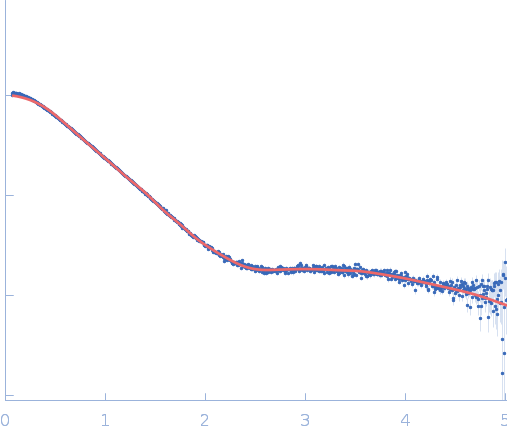
|
| Sample: |
Repeats-in-toxin domain Block V of adenylate cyclase toxin monomer, 19 kDa Bordetella pertussis protein
|
| Buffer: |
50 mM Tris, 5 mM DTT, 10 mM BaCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2024 Aug 1
|
Ion-selective conformational stabilization of a disordered repeats-in-toxin protein domain.
Biophys J (2025)
Gudinas AP, Shambharkar GM, Chang MP, Fernández D, Matsui T, Mai DJ
|
| RgGuinier |
2.7 |
nm |
| Dmax |
11.7 |
nm |
| VolumePorod |
35 |
nm3 |
|
|
|
|
|
|
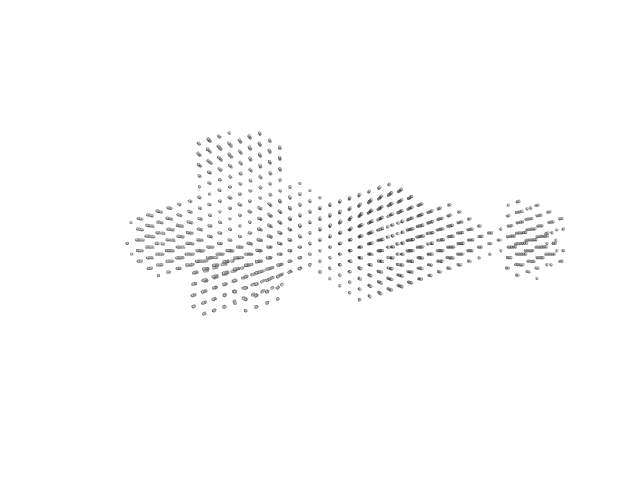
|
| Sample: |
Alpha-hemolysin translocation ATP-binding protein HlyB hexamer, 480 kDa Escherichia coli (strain … protein
HlyA (GFP-Hemolysin with Flag-Tag insertion) monomer, 144 kDa Escherichia coli protein
Membrane fusion protein (MFP) family protein (R14C; L229V) hexamer, 327 kDa Escherichia coli (strain … protein
Outer membrane protein TolC trimer, 161 kDa Escherichia coli (strain … protein
|
| Buffer: |
50 mM Tris, 150 mM NaCl, 10 mM CaCl2, 0.0063% glyco-diosgenin (GDN), pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2023 Jun 29
|
Molecular Insights into CLD Domain Dynamics and Toxin Recruitment of the HlyA E. coli T1SS
Journal of Molecular Biology :169485 (2025)
Gentile R, Schott-Verdugo S, Khosa S, Günes C, Bonus M, Reiners J, Smits S, Schmitt L, Gohlke H
|
| RgGuinier |
11.4 |
nm |
| Dmax |
46.2 |
nm |
| VolumePorod |
2837 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Calcium/calmodulin-dependent protein kinase type II subunit alpha (Δ1-6; Q223K; Δ329-344) dodecamer, 620 kDa Homo sapiens protein
|
| Buffer: |
25 mM tris-HCl pH 8.8, 250 mM KCl, 1 mM TCEP, 2% glycerol, pH: |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2023 Apr 17
|
A domain-swapped CaMKII conformation facilitates linker-mediated allosteric regulation.
Nat Commun 16(1):8461 (2025)
Nguyen BV, Özden C, Dong K, Koc OC, Torres-Ocampo AP, Dziedzic N, Flaherty D, Huang J, Sankara S, Abromson NL, Tomchick DR, Fissore RA, Chen J, Garman SC, Stratton MM
|
| RgGuinier |
6.8 |
nm |
| Dmax |
25.7 |
nm |
| VolumePorod |
1437 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Calcium/calmodulin-dependent protein kinase type II subunit alpha (Δ1-6; Q223K; Δ 316-329) dodecamer, 626 kDa Homo sapiens protein
|
| Buffer: |
25 mM tris-HCl pH 8.8, 250 mM KCl, 1 mM TCEP, 2% glycerol, pH: |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2023 Apr 17
|
A domain-swapped CaMKII conformation facilitates linker-mediated allosteric regulation.
Nat Commun 16(1):8461 (2025)
Nguyen BV, Özden C, Dong K, Koc OC, Torres-Ocampo AP, Dziedzic N, Flaherty D, Huang J, Sankara S, Abromson NL, Tomchick DR, Fissore RA, Chen J, Garman SC, Stratton MM
|
| RgGuinier |
6.0 |
nm |
| Dmax |
19.0 |
nm |
| VolumePorod |
1080 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
GAGE6 60bp dsDNA oligo monomer, 37 kDa Homo sapiens DNA
|
| Buffer: |
150 mM KCl, 20 mM HEPES, 5% glycerol, 5 mM MgCl2, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2023 Mar 14
|
The 60bp GAGE6 dsDNA oligonucleotide
Heidar Koning
|
| RgGuinier |
4.8 |
nm |
| Dmax |
19.5 |
nm |
| VolumePorod |
101 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Murine Immunoglobulin E (IgE) antibodies monomer, 165 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris, 50 mM NaCl, pH: 8.4 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2024 Oct 3
|
Allergen-induced structural rearrangements in IgE: insights from SAXS and molecular dynamics.
Int J Biol Macromol :147658 (2025)
Gómez-Velasco H, García-Ramírez B, Siliqi D, Graewert MA, Quintero-Martinez A, Ortega E, Rodríguez-Romero A
|
| RgGuinier |
5.5 |
nm |
| Dmax |
19.2 |
nm |
| VolumePorod |
422 |
nm3 |
|
|
|
|
|
|
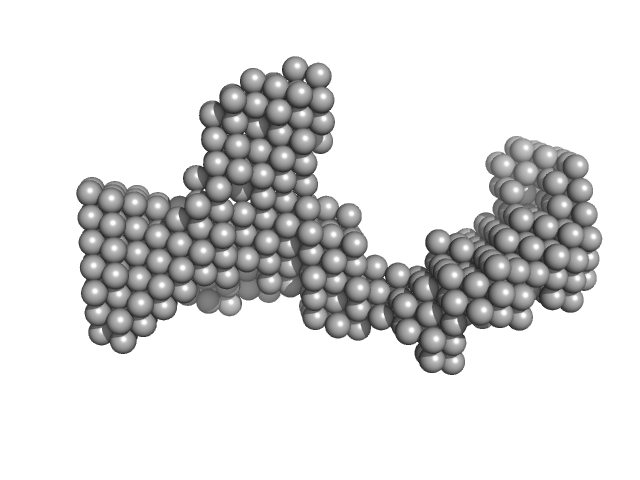
|
| Sample: |
DENV2 5' untranslated terminal region monomer, 57 kDa RNA
|
| Buffer: |
10 mM Bis-Tris, 100 mM NaCl, 5 mM MgCl2, 15 mM KCl, 5% glycerol, pH: 6 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 Sep 10
|
Structural Dynamics of Dengue Virus UTRs and Their Cyclization.
Biophys J (2025)
Robinson ZE, Pereira HS, D'Souza MH, Patel TR
|
| RgGuinier |
3.7 |
nm |
| Dmax |
11.5 |
nm |
| VolumePorod |
57 |
nm3 |
|
|