|
|
|
|

|
| Sample: |
Probable S-adenosyl-L-methionine-dependent RNA methyltransferase RSM22, mitochondrial monomer, 70 kDa Saccharomyces cerevisiae protein
|
| Buffer: |
40 mM Tris pH 7.5, 500 mM NaCl, 5% glycerol, 2.5 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 May 1
|
Probable S-adenosyl-L-methionine-dependent RNA methyltransferase RSM22
Jahangir Alam
|
| RgGuinier |
3.8 |
nm |
| Dmax |
13.9 |
nm |
| VolumePorod |
160 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Dimeric Probable S-adenosyl-L-methionine-dependent RNA methyltransferase RSM22, mitochondrial dimer, 141 kDa Saccharomyces cerevisiae protein
|
| Buffer: |
40 mM Tris pH 7.5, 500 mM NaCl, 5% glycerol, 2.5 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 May 1
|
Probable S-adenosyl-L-methionine-dependent RNA methyltransferase RSM22
Jahangir Alam
|
| RgGuinier |
5.0 |
nm |
| Dmax |
18.2 |
nm |
| VolumePorod |
516 |
nm3 |
|
|
|
|
|
|
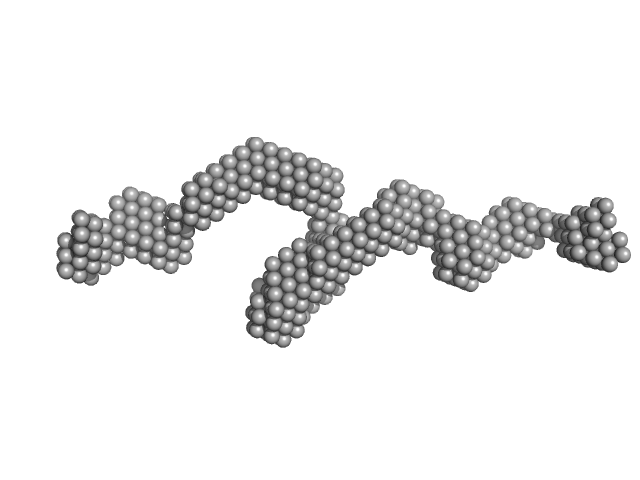
|
| Sample: |
Sperm-associated antigen 1 monomer, 104 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 1% glycerol, 5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Mar 21
|
R2SP
Raphael Dos Santos Morais
|
| RgGuinier |
7.3 |
nm |
| Dmax |
34.0 |
nm |
| VolumePorod |
374 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
RuvB-like 1 hexamer, 311 kDa Homo sapiens protein
RuvB-like 2 hexamer, 311 kDa Homo sapiens protein
Sperm-associated antigen 1 monomer, 104 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 1% glycerol, 5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Mar 21
|
R2SP
Raphael Dos Santos Morais
|
| RgGuinier |
6.8 |
nm |
| Dmax |
34.8 |
nm |
| VolumePorod |
1800 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
RuvB-like 1 hexamer, 311 kDa Homo sapiens protein
RuvB-like 2 hexamer, 311 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 1% glycerol, 5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Mar 3
|
R2SP
Raphael Dos Santos Morais
|
| RgGuinier |
6.3 |
nm |
| Dmax |
23.4 |
nm |
| VolumePorod |
1550 |
nm3 |
|
|
|
|
|
|
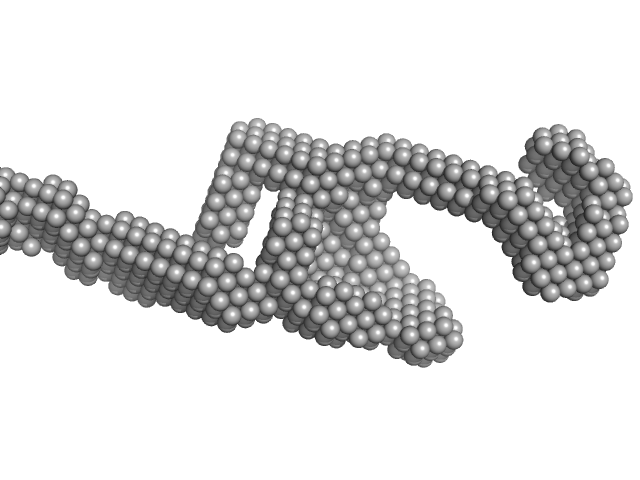
|
| Sample: |
Sperm-associated antigen 1 monomer, 104 kDa Homo sapiens protein
PIH1 domain-containing protein 2 monomer, 36 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 1% glycerol, 5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Mar 21
|
R2SP
Raphael Dos Santos Morais
|
| RgGuinier |
7.0 |
nm |
| Dmax |
30.9 |
nm |
| VolumePorod |
664 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
PIH1 domain-containing protein 2 monomer, 36 kDa Homo sapiens protein
Sperm-associated antigen 1 monomer, 35 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 1% glycerol, 5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Mar 21
|
R2SP
Raphael Dos Santos Morais
|
| RgGuinier |
5.0 |
nm |
| Dmax |
19.3 |
nm |
| VolumePorod |
123 |
nm3 |
|
|
|
|
|
|
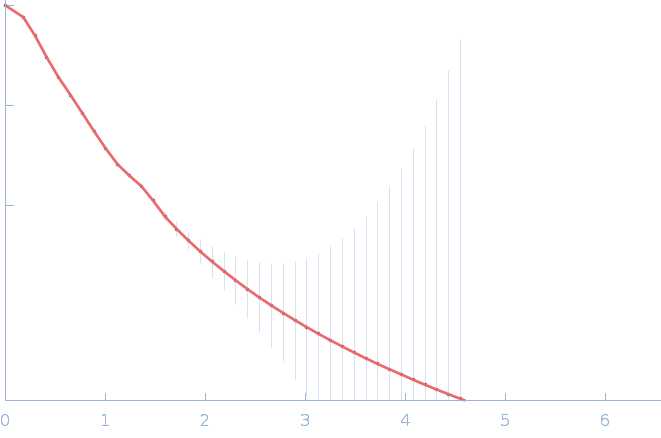
|
| Sample: |
Nitrogen fixation regulatory protein (Q409L) dimer, 115 kDa Azotobacter vinelandii protein
|
| Buffer: |
50 mM Bis-Tris, 100 mM (NH4)2SO4, 10% glycerol, 5 mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2022 Jul 5
|
Structural insights into redox signal transduction mechanisms in the control of nitrogen fixation by the NifLA system
Proceedings of the National Academy of Sciences 120(30) (2023)
Boyer N, Tokmina-Lukaszewska M, Bueno Batista M, Mus F, Dixon R, Bothner B, Peters J
|
| RgGuinier |
4.9 |
nm |
| Dmax |
17.9 |
nm |
| VolumePorod |
233 |
nm3 |
|
|
|
|
|
|
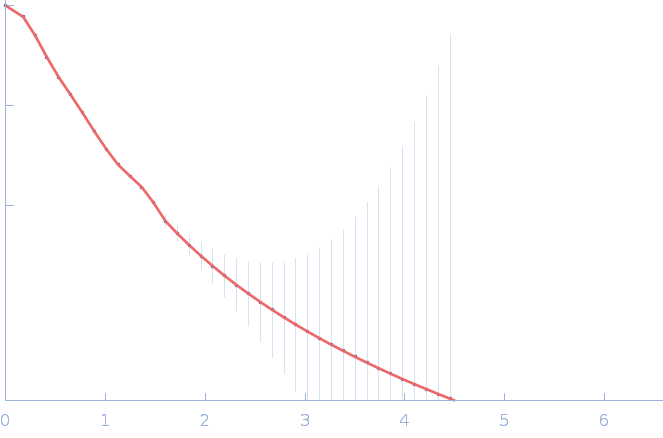
|
| Sample: |
Nitrogen fixation regulatory protein (Q409L) dimer, 115 kDa Azotobacter vinelandii protein
|
| Buffer: |
50 mM Bis-Tris, 100 mM (NH4)2SO4, 10% glycerol, 5 mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2022 Jul 5
|
Structural insights into redox signal transduction mechanisms in the control of nitrogen fixation by the NifLA system
Proceedings of the National Academy of Sciences 120(30) (2023)
Boyer N, Tokmina-Lukaszewska M, Bueno Batista M, Mus F, Dixon R, Bothner B, Peters J
|
| RgGuinier |
4.9 |
nm |
| Dmax |
18.0 |
nm |
| VolumePorod |
225 |
nm3 |
|
|
|
|
|
|
|
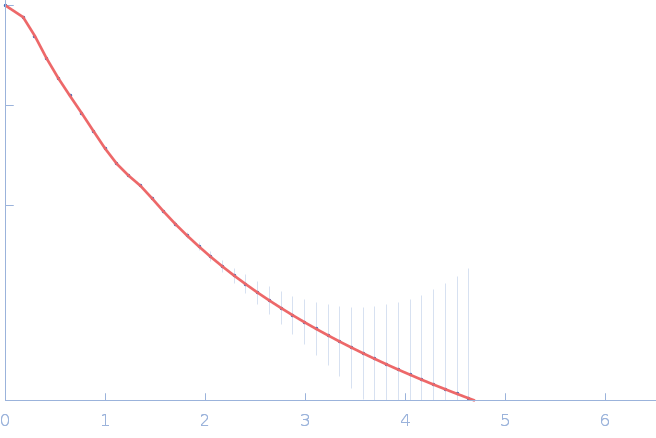
| Sample: |
Nitrogen fixation regulatory protein (Q409L) dimer, 115 kDa Azotobacter vinelandii protein
|
| Buffer: |
50 mM Bis-Tris, 100 mM (NH4)2SO4, 10% glycerol, 5 mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2022 Jul 5
|
Structural insights into redox signal transduction mechanisms in the control of nitrogen fixation by the NifLA system
Proceedings of the National Academy of Sciences 120(30) (2023)
Boyer N, Tokmina-Lukaszewska M, Bueno Batista M, Mus F, Dixon R, Bothner B, Peters J
|
| RgGuinier |
4.9 |
nm |
| Dmax |
18.0 |
nm |
| VolumePorod |
243 |
nm3 |
|
|