|
|
|
|
|
| Sample: |
Nitrogen fixation regulatory protein (Q409L) dimer, 115 kDa Azotobacter vinelandii protein
|
| Buffer: |
50 mM Bis-Tris, 100 mM (NH4)2SO4, 10% glycerol, 5 mM DTT, pH: 7 |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2022 Jul 5
|
Structural insights into redox signal transduction mechanisms in the control of nitrogen fixation by the NifLA system
Proceedings of the National Academy of Sciences 120(30) (2023)
Boyer N, Tokmina-Lukaszewska M, Bueno Batista M, Mus F, Dixon R, Bothner B, Peters J
|
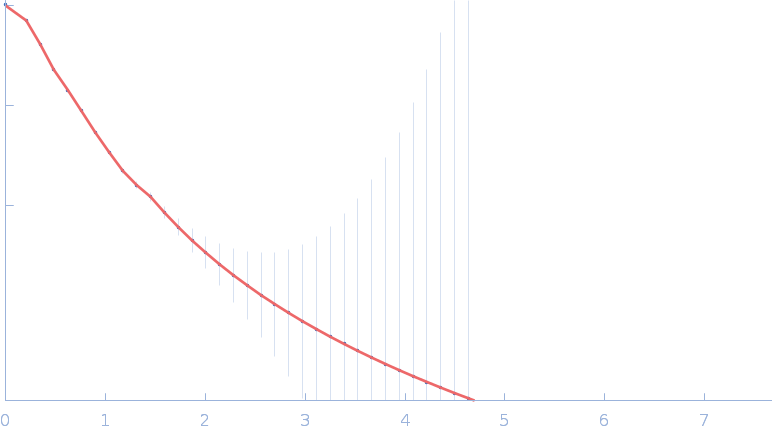
| RgGuinier |
5.1 |
nm |
| Dmax |
17.8 |
nm |
| VolumePorod |
224 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Bartonella effector protein (Bep) substrate of VirB T4SS monomer, 64 kDa Bartonella clarridgeiae (strain … protein
|
| Buffer: |
25 mM Hepes, 300 mM NaCl, 1 mM TCEP, 5% v/v glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at Xenocs Xeuss 2.0 Q-Xoom, Center for Structural Studies, Heinrich-Heine-University on 2020 May 29
|
Structure and function of Bartonella effector protein 1: target and interdomain interactions
University of Basel PhD thesis 15051 (2023)
Markus Huber, Jens Reiners
|
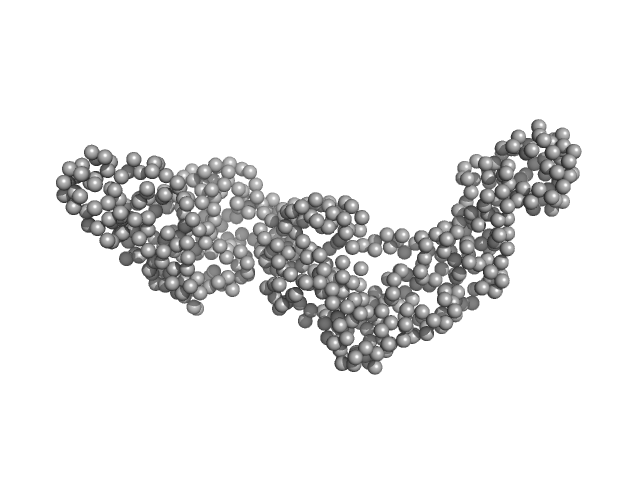
| RgGuinier |
4.1 |
nm |
| Dmax |
13.4 |
nm |
| VolumePorod |
83 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Bartonella effector protein (Bep) substrate of VirB T4SS monomer, 64 kDa Bartonella clarridgeiae (strain … protein
|
| Buffer: |
25 mM Hepes, 300 mM NaCl, 1 mM TCEP, 5% v/v glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Jun 26
|
Structure and function of Bartonella effector protein 1: target and interdomain interactions
University of Basel PhD thesis 15051 (2023)
Markus Huber, Jens Reiners
|
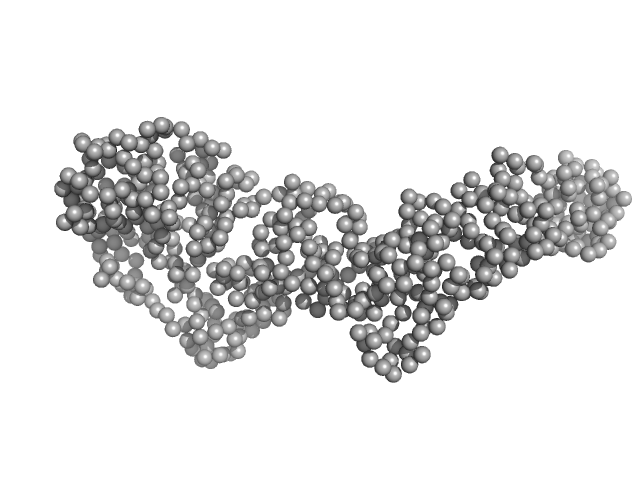
| RgGuinier |
3.8 |
nm |
| Dmax |
13.4 |
nm |
| VolumePorod |
87 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Bartonella effector protein (Bep) substrate of VirB T4SS monomer, 64 kDa Bartonella clarridgeiae (strain … protein
|
| Buffer: |
25 mM Hepes, 300 mM NaCl, 1 mM TCEP, 5% v/v glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Jun 26
|
Structure and function of Bartonella effector protein 1: target and interdomain interactions
University of Basel PhD thesis 15051 (2023)
Markus Huber, Jens Reiners
|
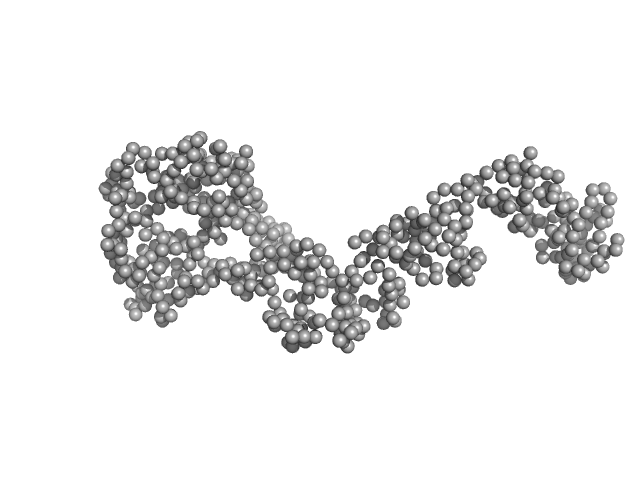
| RgGuinier |
4.1 |
nm |
| Dmax |
14.4 |
nm |
| VolumePorod |
108 |
nm3 |
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDQP8_ensemble_example.png)
|
| Sample: |
Stress response regulating small RNA OxyS monomer, 36 kDa Escherichia coli RNA
|
| Buffer: |
50 mM HEPES pH 6.8, 50 mM NaCl, pH: 6.8 |
| Experiment: |
SAXS
data collected at Austrian SAXS beamline 5.2L, ELETTRA on 2022 Jan 31
|
Spatial arrangement of functional domains in OxyS stress response sRNA
RNA :rna.079618.123 (2023)
Stih V, Amenitsch H, Plavec J, Podbevsek P
|
| RgGuinier |
4.8 |
nm |
| Dmax |
15.5 |
nm |
| VolumePorod |
68 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Guanylate-binding protein 1 monomer, 68 kDa Homo sapiens protein
|
| Buffer: |
50 mM TRIS, 5 mM MgCl2, 150 mM NaCl, pH: 7.9 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Apr 30
|
Integrative dynamic structural biology unveils conformers essential for the oligomerization of a large GTPase.
Elife 12 (2023)
Peulen TO, Hengstenberg CS, Biehl R, Dimura M, Lorenz C, Valeri A, Folz J, Hanke CA, Ince S, Vöpel T, Farago B, Gohlke H, Klare JP, Stadler AM, Seidel CAM, Herrmann C
|
| RgGuinier |
3.9 |
nm |
| Dmax |
14.4 |
nm |
| VolumePorod |
105 |
nm3 |
|
|
|
|
|
|
|
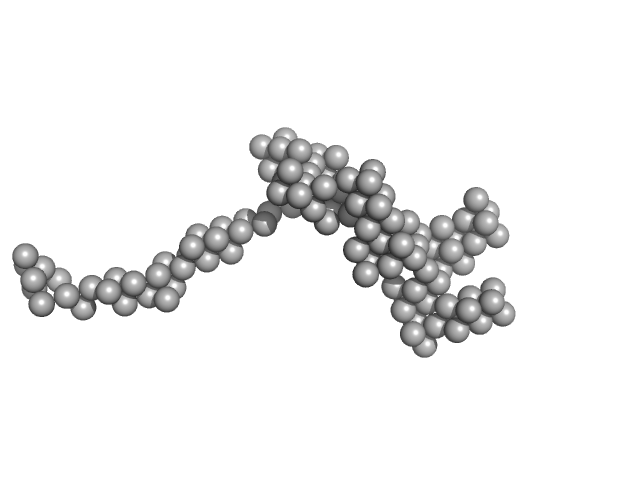
| Sample: |
Receptor-type tyrosine-protein phosphatase alpha dimer, 150 kDa Homo sapiens protein
|
| Buffer: |
10 mM phosphate, 137 mM NaCl, 2.7 mM KCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at 13A, Taiwan Photon Source, NSRRC on 2021 Aug 19
|
High Density of N- and O-Glycosylation Shields and Defines the Structural Dynamics of the Intrinsically Disordered Ectodomain of Receptor-type Protein Tyrosine Phosphatase Alpha
JACS Au (2023)
Chien Y, Wang Y, Sridharan D, Kuo C, Chien C, Uchihashi T, Kato K, Angata T, Meng T, Hsu S, Khoo K
|
| RgGuinier |
8.3 |
nm |
| Dmax |
35.3 |
nm |
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDRN9_fit1_model1.png)
|
| Sample: |
Polysorbate 20 (PS20) with no or low amount of the fatty acid myristic acid (MA) (< 100 µg/ml) 0, 45 kDa
|
| Buffer: |
aqueous solution of 4% methanol, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Nov 2
|
Small-angle x-ray scattering investigation of the integration of free fatty acids in polysorbate 20 micelles
Biophysical Journal (2023)
Ehrit J, Gräwert T, Göddeke H, Konarev P, Svergun D, Nagel N
|
| RgGuinier |
3.4 |
nm |
| Dmax |
8.6 |
nm |
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDRP9_fit1_model1.png)
|
| Sample: |
Polydisperse core-shell ellipsoidal micelles of Polysorbate 20 (PS20) with high amount of the fatty acid myristic acid (MA) (> 500 µg/ml) 0, 45 kDa
|
| Buffer: |
aqueous solution of 4% methanol, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Nov 2
|
Small-angle x-ray scattering investigation of the integration of free fatty acids in polysorbate 20 micelles
Biophysical Journal (2023)
Ehrit J, Gräwert T, Göddeke H, Konarev P, Svergun D, Nagel N
|
| RgGuinier |
3.5 |
nm |
| Dmax |
8.8 |
nm |
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDRQ9_fit1_model1.png)
|
| Sample: |
Polydisperse core-shell ellipsoidal micelles of POE sorbitan monolaurate fraction (F2) with no or low amount of the fatty acid myristic acid (MA) (< 100 µg/ml) 20-mer, 24 kDa
|
| Buffer: |
aqueous solution of 4% methanol, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Mar 6
|
Small-angle x-ray scattering investigation of the integration of free fatty acids in polysorbate 20 micelles
Biophysical Journal (2023)
Ehrit J, Gräwert T, Göddeke H, Konarev P, Svergun D, Nagel N
|
| RgGuinier |
3.0 |
nm |
| Dmax |
8.0 |
nm |
|
|