|
|
|
|
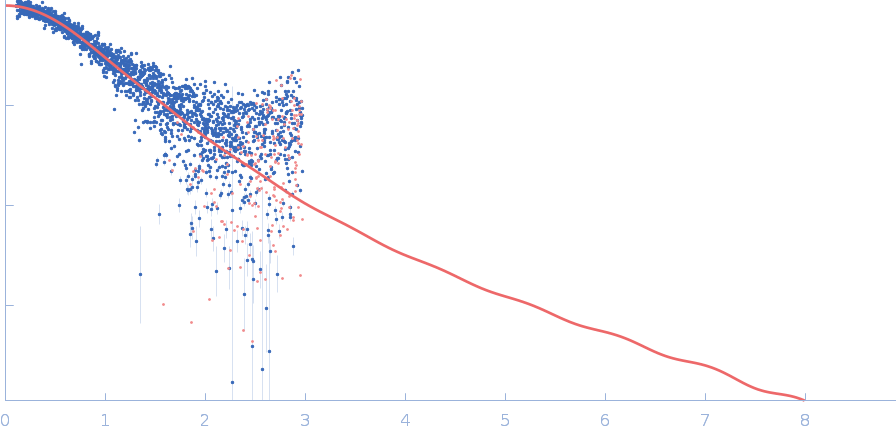
|
| Sample: |
Polyubiquitin-C monomer, 17 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 Feb 1
|
Stretching the chains: the destabilizing impact of Cu(2+) and Zn(2+) ions on K48-linked diubiquitin.
Dalton Trans (2023)
Mangini V, Grasso G, Belviso BD, Sciacca MFM, Lanza V, Caliandro R, Milardi D
|
| RgGuinier |
2.0 |
nm |
| Dmax |
7.5 |
nm |
| VolumePorod |
20 |
nm3 |
|
|
|
|
|
|
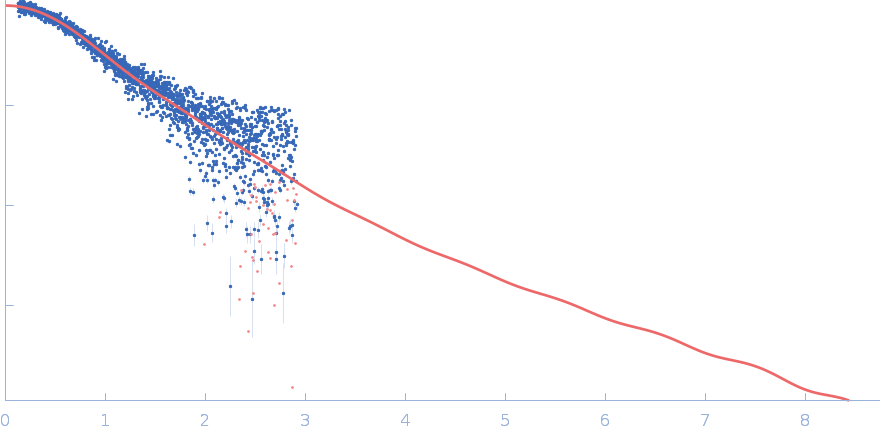
|
| Sample: |
Polyubiquitin-C monomer, 17 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 Feb 1
|
Stretching the chains: the destabilizing impact of Cu(2+) and Zn(2+) ions on K48-linked diubiquitin.
Dalton Trans (2023)
Mangini V, Grasso G, Belviso BD, Sciacca MFM, Lanza V, Caliandro R, Milardi D
|
| RgGuinier |
1.9 |
nm |
| Dmax |
6.3 |
nm |
| VolumePorod |
20 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Kinesin-like protein KIF3B monomer, 32 kDa Mus musculus protein
Kinesin-associated protein 3 monomer, 81 kDa Mus musculus protein
Kinesin-like protein KIF3A monomer, 28 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris-HCl, 200 mM NaCl, 5% glycerol, 1 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at BL-15A2, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2020 Jun 24
|
The two-step cargo recognition mechanism of heterotrimeric kinesin.
EMBO Rep :e56864 (2023)
Jiang X, Ogawa T, Yonezawa K, Shimizu N, Ichinose S, Uchihashi T, Nagaike W, Moriya T, Adachi N, Kawasaki M, Dohmae N, Senda T, Hirokawa N
|
| RgGuinier |
5.8 |
nm |
| Dmax |
27.0 |
nm |
| VolumePorod |
557 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Adenomatous polyposis coli protein (N-terminal ARM domain) monomer, 75 kDa Mus musculus protein
Kinesin-like protein KIF3A monomer, 28 kDa Mus musculus protein
Kinesin-like protein KIF3B monomer, 32 kDa Mus musculus protein
Kinesin-associated protein 3 monomer, 81 kDa Mus musculus protein
|
| Buffer: |
25 mM Tris-HCl, 200 mM NaCl, 1 mM DTT, 5% glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2020 Nov 17
|
The two-step cargo recognition mechanism of heterotrimeric kinesin.
EMBO Rep :e56864 (2023)
Jiang X, Ogawa T, Yonezawa K, Shimizu N, Ichinose S, Uchihashi T, Nagaike W, Moriya T, Adachi N, Kawasaki M, Dohmae N, Senda T, Hirokawa N
|
| RgGuinier |
5.4 |
nm |
| Dmax |
19.5 |
nm |
| VolumePorod |
763 |
nm3 |
|
|
|
|
|
|
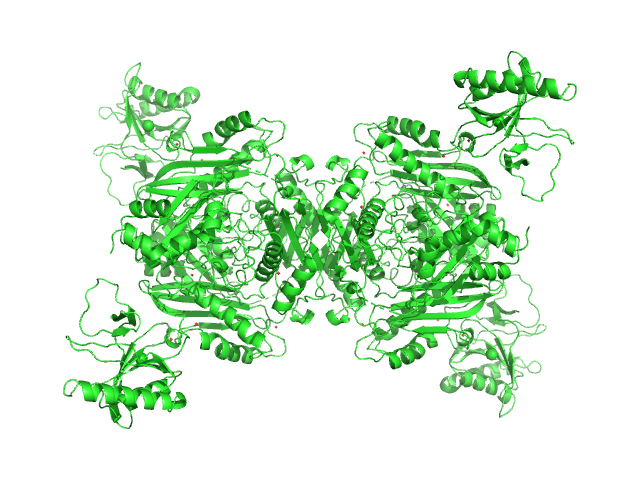
|
| Sample: |
S-adenosylmethionine synthase tetramer, 168 kDa Escherichia coli (strain … protein
S-adenosyl-L-methionine lyase tetramer, 71 kDa Enterobacteria phage T3 protein
|
| Buffer: |
50 mM Tris-HCl, 300 mM NaCl, 5 mM β-mercaptoethanol, 2 % glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2021 Jun 17
|
Phage T3 overcomes the BREX defense through SAM cleavage and inhibition of SAM synthesis by SAM lyase.
Cell Rep 42(8):112972 (2023)
Andriianov A, Trigüis S, Drobiazko A, Sierro N, Ivanov NV, Selmer M, Severinov K, Isaev A
|
| RgGuinier |
4.8 |
nm |
| Dmax |
17.5 |
nm |
| VolumePorod |
325 |
nm3 |
|
|
|
|
|
|
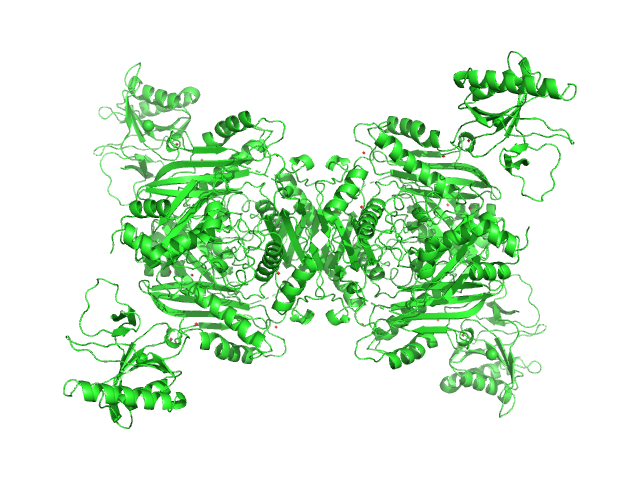
|
| Sample: |
S-adenosylmethionine synthase tetramer, 168 kDa Escherichia coli (strain … protein
S-adenosyl-L-methionine lyase (E68Q/Q94A) tetramer, 71 kDa Enterobacteria phage T3 protein
|
| Buffer: |
50 mM Tris-HCl, 300 mM NaCl, 5 mM β-mercaptoethanol, 2 % glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2021 May 7
|
Phage T3 overcomes the BREX defense through SAM cleavage and inhibition of SAM synthesis by SAM lyase.
Cell Rep 42(8):112972 (2023)
Andriianov A, Trigüis S, Drobiazko A, Sierro N, Ivanov NV, Selmer M, Severinov K, Isaev A
|
| RgGuinier |
4.8 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
326 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Sensor domain-containing diguanylate cyclase dimer, 72 kDa Methylotenera sp. protein
|
| Buffer: |
10 mM Tris, 50 mM NaCl, 2 mM MgCl2, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2023 May 4
|
Illuminating the inner workings of a natural protein switch: Blue-light sensing in LOV-activated diguanylate cyclases
Science Advances 9(31) (2023)
Vide U, Kasapović D, Fuchs M, Heimböck M, Totaro M, Zenzmaier E, Winkler A
|
| RgGuinier |
2.9 |
nm |
| Dmax |
11.1 |
nm |
| VolumePorod |
115 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Pikachurin monomer, 22 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 200 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Mar 3
|
Structure of the photoreceptor synaptic assembly of the extracellular matrix protein pikachurin with the orphan receptor GPR179
Science Signaling 16(795) (2023)
Patil D, Pantalone S, Cao Y, Laboute T, Novick S, Singh S, Savino S, Faravelli S, Magnani F, Griffin P, Singh A, Forneris F, Martemyanov K
|
| RgGuinier |
2.2 |
nm |
| Dmax |
8.2 |
nm |
| VolumePorod |
48 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Pikachurin N-terminal FnIII(1-2) domains monomer, 25 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 200 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Mar 3
|
Structure of the photoreceptor synaptic assembly of the extracellular matrix protein pikachurin with the orphan receptor GPR179
Science Signaling 16(795) (2023)
Patil D, Pantalone S, Cao Y, Laboute T, Novick S, Singh S, Savino S, Faravelli S, Magnani F, Griffin P, Singh A, Forneris F, Martemyanov K
|
| RgGuinier |
3.4 |
nm |
| Dmax |
16.4 |
nm |
| VolumePorod |
44 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Pikachurin monomer, 26 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 200 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Mar 3
|
Structure of the photoreceptor synaptic assembly of the extracellular matrix protein pikachurin with the orphan receptor GPR179
Science Signaling 16(795) (2023)
Patil D, Pantalone S, Cao Y, Laboute T, Novick S, Singh S, Savino S, Faravelli S, Magnani F, Griffin P, Singh A, Forneris F, Martemyanov K
|
| RgGuinier |
2.3 |
nm |
| Dmax |
7.4 |
nm |
| VolumePorod |
52 |
nm3 |
|
|