|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDRR9_fit1_model1.png)
|
| Sample: |
Polydisperse core-shell ellipsoidal micelles of POE sorbitan monolaurate fraction (F2) with high amount of the fatty acid myristic acid (MA) (> 500 µg/ml) 22-mer, 26 kDa
|
| Buffer: |
aqueous solution of 4% methanol, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Mar 6
|
Small-angle x-ray scattering investigation of the integration of free fatty acids in polysorbate 20 micelles
Biophysical Journal (2023)
Ehrit J, Gräwert T, Göddeke H, Konarev P, Svergun D, Nagel N
|
| RgGuinier |
3.1 |
nm |
| Dmax |
8.3 |
nm |
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDRS9_fit1_model1.png)
|
| Sample: |
Polydisperse core-shell ellipsoidal micelles of POE sorbitan higher order esters fraction (F4) with no or low amount of the fatty acid myristic acid (MA) (< 100 µg/ml) 0, 42 kDa
|
| Buffer: |
aqueous solution of 4% methanol, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Oct 29
|
Small-angle x-ray scattering investigation of the integration of free fatty acids in polysorbate 20 micelles
Biophysical Journal (2023)
Ehrit J, Gräwert T, Göddeke H, Konarev P, Svergun D, Nagel N
|
| RgGuinier |
3.3 |
nm |
| Dmax |
8.3 |
nm |
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDRT9_fit1_model1.png)
|
| Sample: |
Polydisperse core-shell ellipsoidal micelles of POE sorbitan higher order esters fraction (F4) with high amount of the fatty acid myristic acid (MA) (> 500 µg/ml) 0, 42 kDa
|
| Buffer: |
aqueous solution of 4% methanol, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Oct 29
|
Small-angle x-ray scattering investigation of the integration of free fatty acids in polysorbate 20 micelles
Biophysical Journal (2023)
Ehrit J, Gräwert T, Göddeke H, Konarev P, Svergun D, Nagel N
|
| RgGuinier |
3.5 |
nm |
| Dmax |
8.7 |
nm |
|
|
|
|
|
|

|
| Sample: |
At3g55760 monomer, 60 kDa Arabidopsis thaliana protein
|
| Buffer: |
50 mM Tris, 150 mM NaCl, 10% glycerol, 2 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2016 Dec 8
|
LIKE EARLY STARVATION 1 and EARLY STARVATION 1 promote and stabilize amylopectin phase transition in starch biosynthesis.
Sci Adv 9(21):eadg7448 (2023)
Liu C, Pfister B, Osman R, Ritter M, Heutinck A, Sharma M, Eicke S, Fischer-Stettler M, Seung D, Bompard C, Abt MR, Zeeman SC
|
| RgGuinier |
3.7 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
121 |
nm3 |
|
|
|
|
|
|
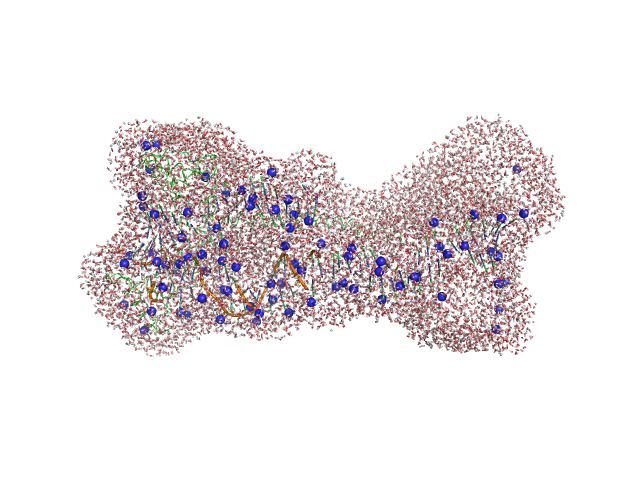
|
| Sample: |
Gcf1p monomer, 26 kDa Candida albicans (strain … protein
Af2_20 DNA monomer, 12 kDa DNA
|
| Buffer: |
25 mM Tris, 20 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Aug 30
|
Structural analysis of the Candida albicans mitochondrial DNA maintenance factor Gcf1p reveals a dynamic DNA-bridging mechanism.
Nucleic Acids Res (2023)
Tarrés-Solé A, Battistini F, Gerhold JM, Piétrement O, Martínez-García B, Ruiz-López E, Lyonnais S, Bernadó P, Roca J, Orozco M, Le Cam E, Sedman J, Solà M
|
| RgGuinier |
3.6 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
92 |
nm3 |
|
|
|
|
|
|
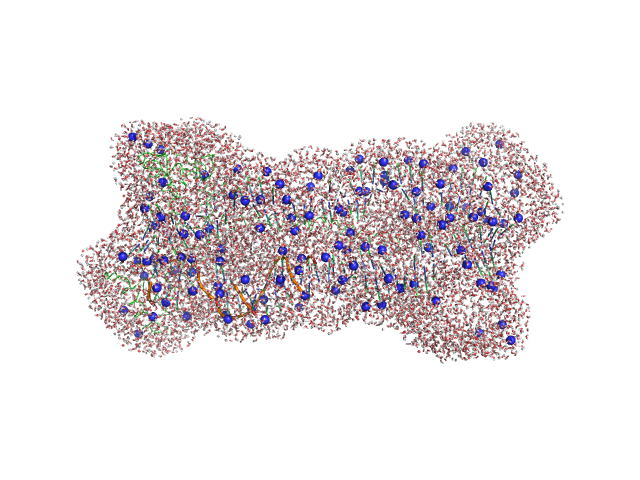
|
| Sample: |
Gcf1p monomer, 26 kDa Candida albicans (strain … protein
Af2_20 DNA monomer, 12 kDa DNA
|
| Buffer: |
25 mM Tris, 20 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Aug 30
|
Structural analysis of the Candida albicans mitochondrial DNA maintenance factor Gcf1p reveals a dynamic DNA-bridging mechanism.
Nucleic Acids Res (2023)
Tarrés-Solé A, Battistini F, Gerhold JM, Piétrement O, Martínez-García B, Ruiz-López E, Lyonnais S, Bernadó P, Roca J, Orozco M, Le Cam E, Sedman J, Solà M
|
| RgGuinier |
3.9 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
133 |
nm3 |
|
|
|
|
|
|
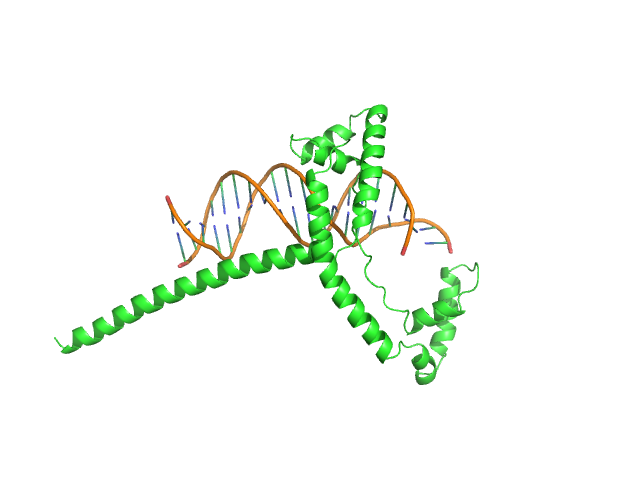
|
| Sample: |
Af2_20 DNA monomer, 12 kDa DNA
Gcf1p(Δ58) monomer, 22 kDa Candida albicans (strain … protein
|
| Buffer: |
25 mM Tris, 20 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Aug 30
|
Structural analysis of the Candida albicans mitochondrial DNA maintenance factor Gcf1p reveals a dynamic DNA-bridging mechanism.
Nucleic Acids Res (2023)
Tarrés-Solé A, Battistini F, Gerhold JM, Piétrement O, Martínez-García B, Ruiz-López E, Lyonnais S, Bernadó P, Roca J, Orozco M, Le Cam E, Sedman J, Solà M
|
| RgGuinier |
2.6 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
52 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Monekypox DNA sequence 1 monomer, 6 kDa Monkeypox virus DNA
|
| Buffer: |
20 mM HEPES, 100 mM KCl, and 0.2 mM EDTA, pH: 7.4 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 Sep 23
|
Mapping and characterization of G‐quadruplexes in monkeypox genomes
Journal of Medical Virology 95(5) (2023)
Pereira H, Gemmill D, Siddiqui M, Vasudeva G, Patel T
|
| RgGuinier |
1.7 |
nm |
| Dmax |
4.4 |
nm |
| VolumePorod |
14 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Monekypox DNA sequence 1 monomer, 6 kDa Monkeypox virus DNA
|
| Buffer: |
20 mM HEPES, 100 mM KCl, and 0.2 mM EDTA, pH: 7.4 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 Sep 23
|
Mapping and characterization of G‐quadruplexes in monkeypox genomes
Journal of Medical Virology 95(5) (2023)
Pereira H, Gemmill D, Siddiqui M, Vasudeva G, Patel T
|
| RgGuinier |
1.6 |
nm |
| Dmax |
4.5 |
nm |
| VolumePorod |
11 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Monekypox DNA sequence 1 mutant monomer, 6 kDa Monkeypox virus DNA
|
| Buffer: |
20 mM HEPES, 100 mM KCl, and 0.2 mM EDTA, pH: 7.4 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 Sep 23
|
Mapping and characterization of G‐quadruplexes in monkeypox genomes
Journal of Medical Virology 95(5) (2023)
Pereira H, Gemmill D, Siddiqui M, Vasudeva G, Patel T
|
| RgGuinier |
1.9 |
nm |
| Dmax |
5.5 |
nm |
| VolumePorod |
16 |
nm3 |
|
|