|
|
|
|
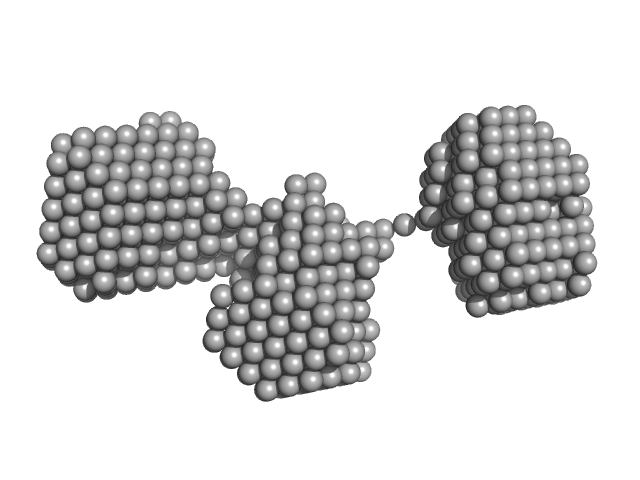
|
| Sample: |
Monekypox DNA sequence 2 mutant monomer, 6 kDa Monkeypox virus DNA
|
| Buffer: |
20 mM HEPES, 100 mM KCl, and 0.2 mM EDTA, pH: 7.4 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 Sep 23
|
Mapping and characterization of G‐quadruplexes in monkeypox genomes
Journal of Medical Virology 95(5) (2023)
Pereira H, Gemmill D, Siddiqui M, Vasudeva G, Patel T
|
| RgGuinier |
1.9 |
nm |
| Dmax |
6.0 |
nm |
| VolumePorod |
8 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
TnpA transposase dimer, 234 kDa Bacillus thuringiensis serovar … protein
|
| Buffer: |
50 mM HEPES, 200 mM NaCl, 100 mM L-Arg HCL, pH: 7.9 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2017 Nov 2
|
AFM-based force spectroscopy unravels stepwise formation of the DNA transposition complex in the widespread Tn3 family mobile genetic elements.
Nucleic Acids Res (2023)
Fernandez M, Shkumatov AV, Liu Y, Stulemeijer C, Derclaye S, Efremov RG, Hallet B, Alsteens D
|
| RgGuinier |
4.6 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
480 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Cell division protein FtsB hexamer, 70 kDa Escherichia coli (strain … protein
Cell division protein FtsL hexamer, 82 kDa Escherichia coli (strain … protein
Cell division protein FtsQ hexamer, 189 kDa Escherichia coli (strain … protein
|
| Buffer: |
20 mM HEPES pH 7.5, 200 mM NaCl, 0.2% of Cymal-5, pH: 7.5 |
| Experiment: |
SAXS
data collected at 13A, Taiwan Photon Source, NSRRC on 2022 Sep 23
|
Structure of the heterotrimeric membrane protein complex FtsB-FtsL-FtsQ of the bacterial divisome
Nature Communications 14(1) (2023)
Nguyen H, Chen X, Parada C, Luo A, Shih O, Jeng U, Huang C, Shih Y, Ma C
|
| RgGuinier |
5.3 |
nm |
| Dmax |
16.5 |
nm |
| VolumePorod |
421 |
nm3 |
|
|
|
|
|
|
|
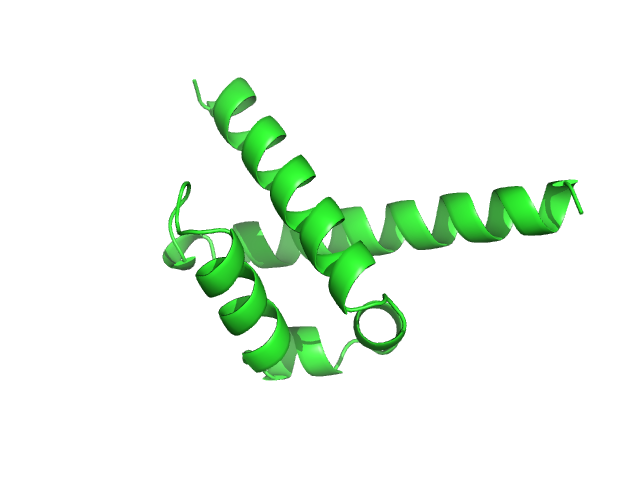
| Sample: |
Pre-mRNA-processing factor 40 homolog A monomer, 9 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 100 mM NaCl, 2 mM CaCl2, 1 mM TCEP, pH: 7.4 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2021 Sep 23
|
Binding by calmodulin is coupled to transient unfolding of the third FF domain of Prp40A.
Protein Sci 32(4):e4606 (2023)
Díaz Casas A, Cordoba JJ, Ferrer BJ, Balakrishnan S, Wurm JE, Pastrana-Ríos B, Chazin WJ
|
| RgGuinier |
1.6 |
nm |
| Dmax |
7.2 |
nm |
| VolumePorod |
13 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Japanese encephaltitis 5' TR monomer, 74 kDa Japanese encephalitis virus RNA
|
| Buffer: |
10 mM Bis-tris, 100 mM NaCl, 15 mM KCl 15 mM MgCl2, 10% glycerol, pH: 5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 May 22
|
Investigating RNA-RNA interactions through computational and biophysical analysis.
Nucleic Acids Res (2023)
Mrozowich T, Park SM, Waldl M, Henrickson A, Tersteeg S, Nelson CR, De Klerk A, Demeler B, Hofacker IL, Wolfinger MT, Patel TR
|
| RgGuinier |
7.0 |
nm |
| Dmax |
22.4 |
nm |
| VolumePorod |
356 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Japanese encephalitis virus 3' UTR monomer, 186 kDa Japanese encephalitis virus RNA
|
| Buffer: |
10 mM Bis-tris, 100 mM NaCl, 15 mM KCl 15 mM MgCl2, 10% glycerol, pH: 5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 May 22
|
Investigating RNA-RNA interactions through computational and biophysical analysis.
Nucleic Acids Res (2023)
Mrozowich T, Park SM, Waldl M, Henrickson A, Tersteeg S, Nelson CR, De Klerk A, Demeler B, Hofacker IL, Wolfinger MT, Patel TR
|
| RgGuinier |
11.3 |
nm |
| Dmax |
34.6 |
nm |
| VolumePorod |
2900 |
nm3 |
|
|
|
|
|
|
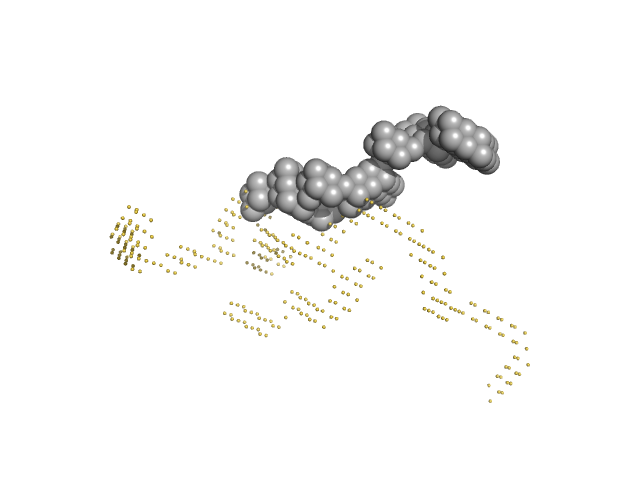
|
| Sample: |
Japanese encephalitis virus 5' TR and 3' UTR complex monomer, 260 kDa Japanese encephalitis virus RNA
|
| Buffer: |
10 mM Bis-tris, 100 mM NaCl, 15 mM KCl 15 mM MgCl2, 10% glycerol, pH: 5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 May 22
|
Investigating RNA-RNA interactions through computational and biophysical analysis.
Nucleic Acids Res (2023)
Mrozowich T, Park SM, Waldl M, Henrickson A, Tersteeg S, Nelson CR, De Klerk A, Demeler B, Hofacker IL, Wolfinger MT, Patel TR
|
| RgGuinier |
12.8 |
nm |
| Dmax |
40.0 |
nm |
| VolumePorod |
4550 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Cell wall assembly regulator SMI1 monomer, 31 kDa Saccharomyces cerevisiae (strain … protein
|
| Buffer: |
100 mM MES, 300 mM NaCl, pH: 6 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Mar 18
|
The conserved yeast protein Knr4 involved in cell wall integrity is a multi-domain intrinsically disordered protein.
J Mol Biol :168048 (2023)
Batista M, Donker EIM, Bon C, Guillien M, Caisso A, Mourey Funding L, Marie François Funding J, Maveyraud L, Zerbib D
|
| RgGuinier |
2.3 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
62 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Cell wall assembly regulator SMI1 monomer, 40 kDa Saccharomyces cerevisiae (strain … protein
|
| Buffer: |
100 mM MES, 300 mM NaCl, pH: 6 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Mar 4
|
The conserved yeast protein Knr4 involved in cell wall integrity is a multi-domain intrinsically disordered protein.
J Mol Biol :168048 (2023)
Batista M, Donker EIM, Bon C, Guillien M, Caisso A, Mourey Funding L, Marie François Funding J, Maveyraud L, Zerbib D
|
| RgGuinier |
2.8 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
79 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Cell wall assembly regulator SMI1 monomer, 57 kDa Saccharomyces cerevisiae (strain … protein
|
| Buffer: |
100 mM MES, 300 mM NaCl, pH: 6 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Mar 7
|
The conserved yeast protein Knr4 involved in cell wall integrity is a multi-domain intrinsically disordered protein.
J Mol Biol :168048 (2023)
Batista M, Donker EIM, Bon C, Guillien M, Caisso A, Mourey Funding L, Marie François Funding J, Maveyraud L, Zerbib D
|
| RgGuinier |
4.8 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
117 |
nm3 |
|
|