|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDQR8_fit1_model1.png)
|
| Sample: |
Huntingtin monomer, 39 kDa Homo sapiens protein
|
| Buffer: |
20mM BisTris-HCl, 150mM NaCl, pH: 6.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Dec 3
|
The structure of pathogenic huntingtin exon 1 defines the bases of its aggregation propensity.
Nat Struct Mol Biol (2023)
Elena-Real CA, Sagar A, Urbanek A, Popovic M, Morató A, Estaña A, Fournet A, Doucet C, Lund XL, Shi ZD, Costa L, Thureau A, Allemand F, Swenson RE, Milhiet PE, Crehuet R, Barducci A, Cortés J, Sinnaeve D, Sibille N, Bernadó P
|
| RgGuinier |
3.3 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
63 |
nm3 |
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDQS8_fit1_model1.png)
|
| Sample: |
Huntingtin monomer, 43 kDa Homo sapiens protein
|
| Buffer: |
20mM BisTris-HCl, 150mM NaCl, pH: 6.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Oct 28
|
The structure of pathogenic huntingtin exon 1 defines the bases of its aggregation propensity.
Nat Struct Mol Biol (2023)
Elena-Real CA, Sagar A, Urbanek A, Popovic M, Morató A, Estaña A, Fournet A, Doucet C, Lund XL, Shi ZD, Costa L, Thureau A, Allemand F, Swenson RE, Milhiet PE, Crehuet R, Barducci A, Cortés J, Sinnaeve D, Sibille N, Bernadó P
|
|
|
|
|
|
|
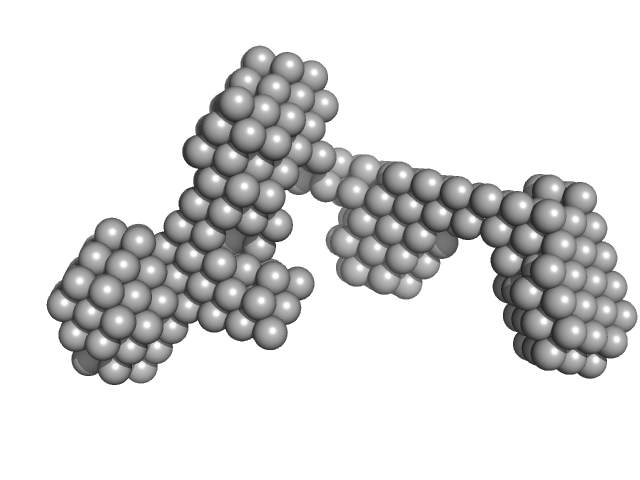
|
| Sample: |
Sal-like protein 4 monomer, 8 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris-HCl, pH 7.5, 200 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2020 Feb 6
|
Structure of SALL4 zinc finger domain reveals link between AT-rich DNA binding and Okihiro syndrome.
Life Sci Alliance 6(3) (2023)
Watson JA, Pantier R, Jayachandran U, Chhatbar K, Alexander-Howden B, Kruusvee V, Prendecki M, Bird A, Cook AG
|
| RgGuinier |
2.1 |
nm |
| Dmax |
5.8 |
nm |
| VolumePorod |
11 |
nm3 |
|
|
|
|
|
|
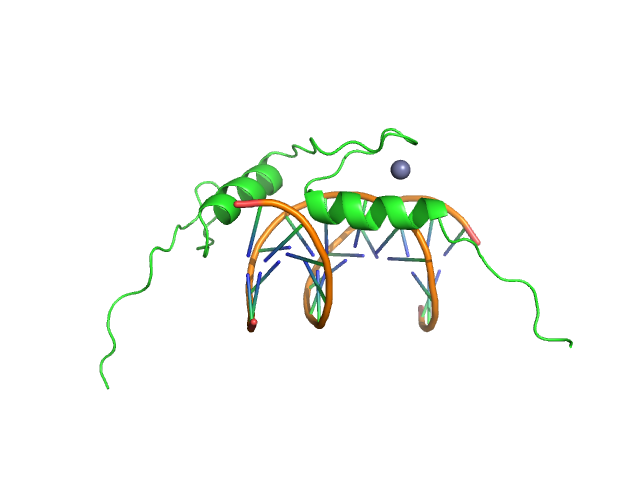
|
| Sample: |
Sal-like protein 4 monomer, 8 kDa Mus musculus protein
AT-rich dsDNA monomer, 7 kDa DNA
|
| Buffer: |
20 mM Tris-HCl, pH 7.5, 200 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2020 Feb 6
|
Structure of SALL4 zinc finger domain reveals link between AT-rich DNA binding and Okihiro syndrome.
Life Sci Alliance 6(3) (2023)
Watson JA, Pantier R, Jayachandran U, Chhatbar K, Alexander-Howden B, Kruusvee V, Prendecki M, Bird A, Cook AG
|
| RgGuinier |
1.8 |
nm |
| Dmax |
5.1 |
nm |
| VolumePorod |
14 |
nm3 |
|
|
|
|
|
|
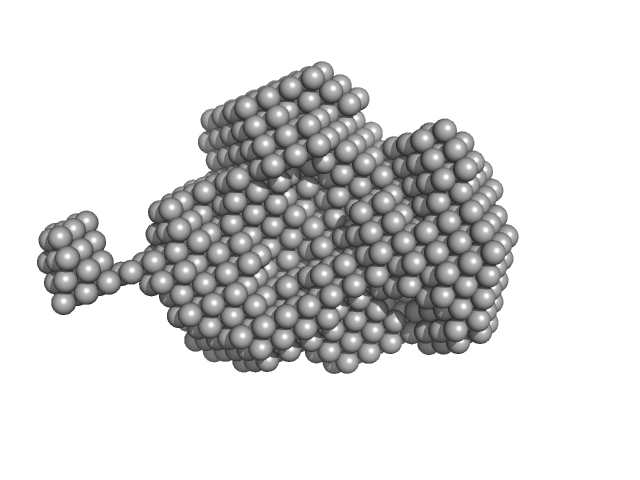
|
| Sample: |
Nucleoside triphosphate pyrophosphohydrolase dimer, 41 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
20 mM Tris-HCl, 100 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2020 Dec 11
|
Structural analysis of the housecleaning nucleoside triphosphate pyrophosphohydrolase MazG from Mycobacterium tuberculosis
Frontiers in Microbiology 14 (2023)
Wang S, Gao B, Chen A, Zhang Z, Wang S, Lv L, Zhao G, Li J
|
| RgGuinier |
2.7 |
nm |
| Dmax |
8.4 |
nm |
| VolumePorod |
64 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Nucleoside triphosphate pyrophosphohydrolase dimer, 71 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2022 Sep 4
|
Structural analysis of the housecleaning nucleoside triphosphate pyrophosphohydrolase MazG from Mycobacterium tuberculosis
Frontiers in Microbiology 14 (2023)
Wang S, Gao B, Chen A, Zhang Z, Wang S, Lv L, Zhao G, Li J
|
| RgGuinier |
3.4 |
nm |
| Dmax |
12.2 |
nm |
| VolumePorod |
121 |
nm3 |
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDQB9_fit1_model1.png)
|
| Sample: |
YdaT_toxin domain-containing protein tetramer, 74 kDa Escherichia coli O157:H7 protein
|
| Buffer: |
20 mM Tris-HCl, 200 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2022 Apr 16
|
Structural basis of DNA binding by YdaT, a functional equivalent of the CII repressor in the cryptic prophage CP-933P from Escherichia coli
O157:H7
Acta Crystallographica Section D Structural Biology 79(3):245-258 (2023)
Prolič-Kalinšek M, Volkov A, Hadži S, Van Dyck J, Bervoets I, Charlier D, Loris R
|
| RgGuinier |
3.5 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
130 |
nm3 |
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDQC9_fit1_model1.png)
|
| Sample: |
YdaT_toxin domain-containing protein (mutant: L111N, F118R) monomer, 18 kDa Escherichia coli O157:H7 protein
|
| Buffer: |
20 mM Tris-HCl, 200 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2021 Apr 14
|
Structural basis of DNA binding by YdaT, a functional equivalent of the CII repressor in the cryptic prophage CP-933P from Escherichia coli
O157:H7
Acta Crystallographica Section D Structural Biology 79(3):245-258 (2023)
Prolič-Kalinšek M, Volkov A, Hadži S, Van Dyck J, Bervoets I, Charlier D, Loris R
|
| RgGuinier |
2.4 |
nm |
| Dmax |
8.2 |
nm |
| VolumePorod |
32 |
nm3 |
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDQD9_fit1_model1.png)
|
| Sample: |
YdaT_toxin domain-containing protein monomer, 13 kDa Escherichia coli O157:H7 protein
|
| Buffer: |
20 mM Tris-HCl, 200 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2022 Apr 16
|
Structural basis of DNA binding by YdaT, a functional equivalent of the CII repressor in the cryptic prophage CP-933P from Escherichia coli
O157:H7
Acta Crystallographica Section D Structural Biology 79(3):245-258 (2023)
Prolič-Kalinšek M, Volkov A, Hadži S, Van Dyck J, Bervoets I, Charlier D, Loris R
|
| RgGuinier |
1.7 |
nm |
| Dmax |
5.7 |
nm |
| VolumePorod |
29 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Iron-utilization periplasmic protein monomer, 34 kDa Haemophilus influenzae protein
|
| Buffer: |
1 mM Na2HPO4.7H2O, 0.18 mM KH2PO4, 13.7 mM NaCl, 0.27 mM KCl, 5%v/v Glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Aug 20
|
Conformational multiplicity of bacterial ferric binding protein revealed by small angle x-ray scattering and molecular dynamics calculations
The Journal of Chemical Physics 158(8) (2023)
Liu G, Ekmen E, Jalalypour F, Mertens H, Jeffries C, Svergun D, Atilgan A, Atilgan C, Sayers Z
|
| RgGuinier |
2.1 |
nm |
| Dmax |
6.3 |
nm |
| VolumePorod |
28 |
nm3 |
|
|