|
|
|
|
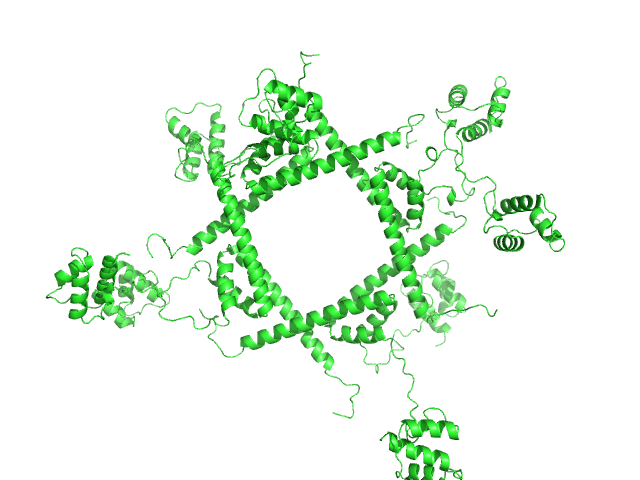
|
| Sample: |
Phage repressor protein CI (C120S) octamer, 122 kDa Escherichia coli O157:H7 protein
|
| Buffer: |
20 mM Tris-HCl, 200 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2022 Apr 16
|
Regulation of the Escherichia coli paaR2-paaA2-ParD2 toxin-antitoxin system
Marusa Prolic Kalinsek
|
| RgGuinier |
5.0 |
nm |
| Dmax |
16.8 |
nm |
| VolumePorod |
428 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Emfourin (Protealysin-associated protein) monomer, 13 kDa Serratia proteamaculans protein
|
| Buffer: |
50 mM Tris-HCL, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Apr 22
|
NMR structure of emfourin, a novel protein metalloprotease inhibitor: insights into the mechanism of action.
J Biol Chem :104585 (2023)
Bozin T, Berdyshev I, Chukhontseva K, Karaseva M, Konarev P, Varizhuk A, Lesovoy D, Arseniev A, Kostrov S, Bocharov E, Demidyuk I
|
| RgGuinier |
1.7 |
nm |
| Dmax |
5.5 |
nm |
| VolumePorod |
16 |
nm3 |
|
|
|
|
|
|
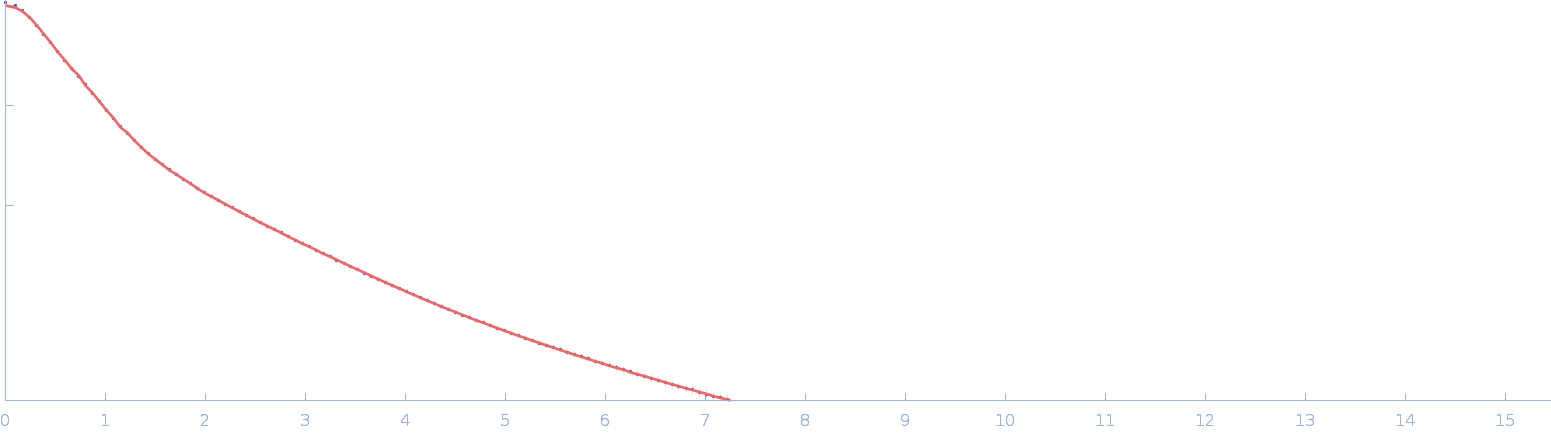
|
| Sample: |
Netrin-1 monomer, 50 kDa Gallus gallus protein
|
| Buffer: |
20 mM Tris-HCl, 500 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Oct 24
|
The dynamic nature of netrin-1 and the structural basis for glycosaminoglycan fragment-induced filament formation
Nature Communications 14(1) (2023)
Meier M, Gupta M, Akgül S, McDougall M, Imhof T, Nikodemus D, Reuten R, Moya-Torres A, To V, Ferens F, Heide F, Padilla-Meier G, Kukura P, Huang W, Gerisch B, Mörgelin M, Poole K, Antebi A, Koch M, Stetefeld J
|
| RgGuinier |
4.0 |
nm |
| Dmax |
18.0 |
nm |
| VolumePorod |
103 |
nm3 |
|
|
|
|
|
|
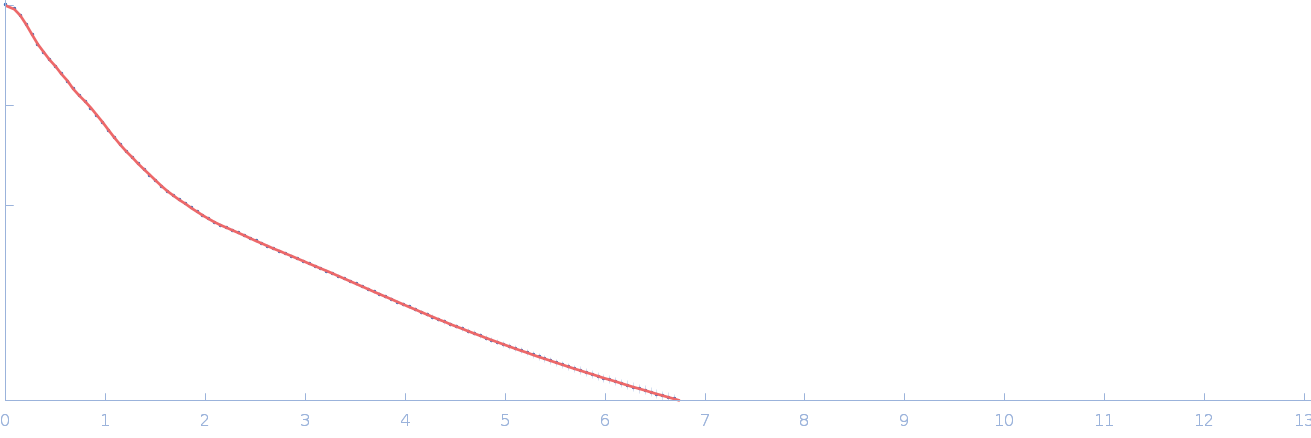
|
| Sample: |
Netrin-1 dimer, 99 kDa Gallus gallus protein
|
| Buffer: |
20 mM Tris-HCl, 500 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Oct 24
|
The dynamic nature of netrin-1 and the structural basis for glycosaminoglycan fragment-induced filament formation
Nature Communications 14(1) (2023)
Meier M, Gupta M, Akgül S, McDougall M, Imhof T, Nikodemus D, Reuten R, Moya-Torres A, To V, Ferens F, Heide F, Padilla-Meier G, Kukura P, Huang W, Gerisch B, Mörgelin M, Poole K, Antebi A, Koch M, Stetefeld J
|
| RgGuinier |
5.9 |
nm |
| Dmax |
21.3 |
nm |
| VolumePorod |
145 |
nm3 |
|
|
|
|
|
|
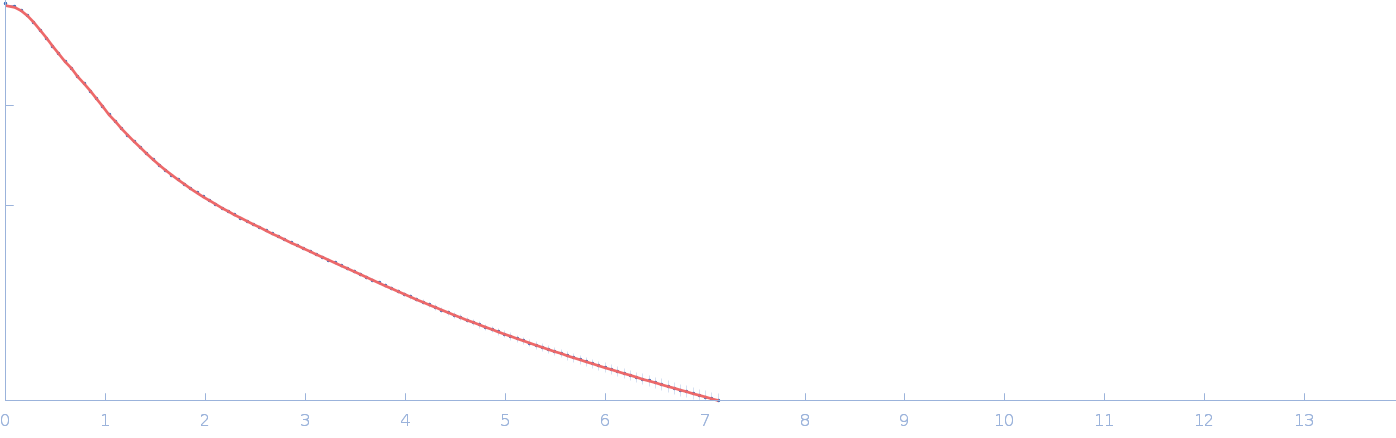
|
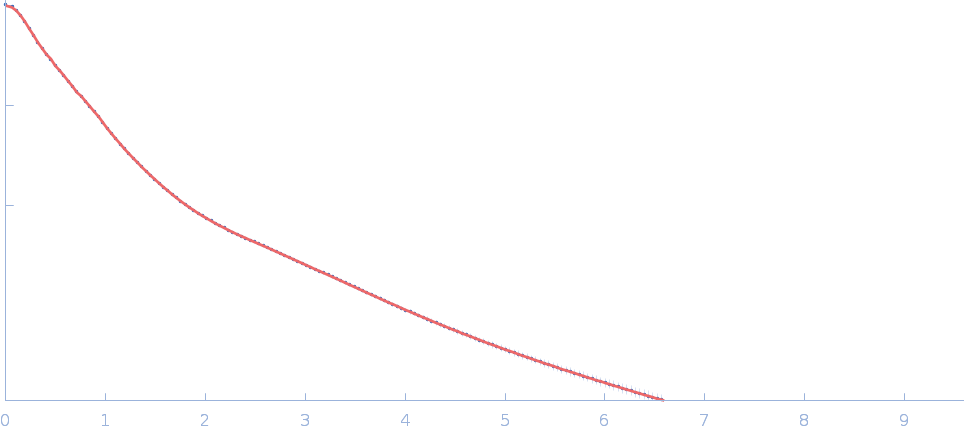
| Sample: |
Netrin-1 monomer, 50 kDa Gallus gallus protein
|
| Buffer: |
50 mM Tris-HCl, 200 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Oct 24
|
The dynamic nature of netrin-1 and the structural basis for glycosaminoglycan fragment-induced filament formation
Nature Communications 14(1) (2023)
Meier M, Gupta M, Akgül S, McDougall M, Imhof T, Nikodemus D, Reuten R, Moya-Torres A, To V, Ferens F, Heide F, Padilla-Meier G, Kukura P, Huang W, Gerisch B, Mörgelin M, Poole K, Antebi A, Koch M, Stetefeld J
|
| RgGuinier |
4.1 |
nm |
| Dmax |
21.0 |
nm |
| VolumePorod |
84 |
nm3 |
|
|
|
|
|
|
|
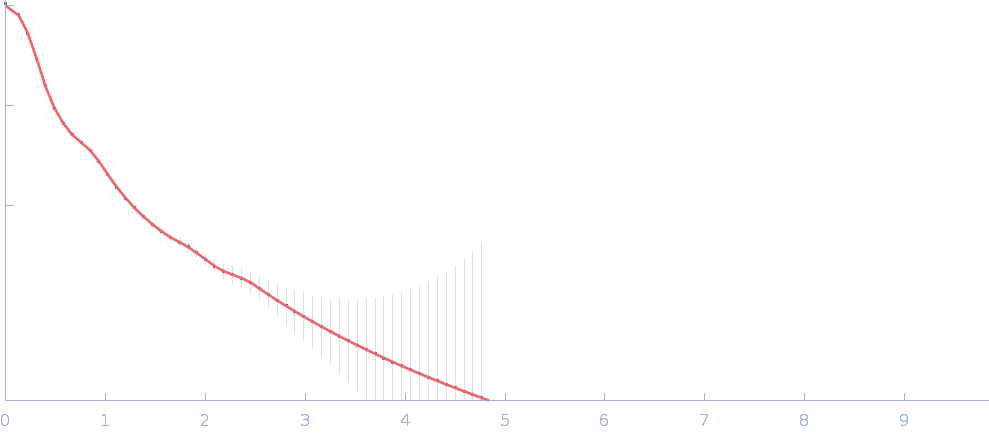
| Sample: |
Netrin-1 dimer, 99 kDa Gallus gallus protein
|
| Buffer: |
50 mM Tris-HCl, 200 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Oct 24
|
The dynamic nature of netrin-1 and the structural basis for glycosaminoglycan fragment-induced filament formation
Nature Communications 14(1) (2023)
Meier M, Gupta M, Akgül S, McDougall M, Imhof T, Nikodemus D, Reuten R, Moya-Torres A, To V, Ferens F, Heide F, Padilla-Meier G, Kukura P, Huang W, Gerisch B, Mörgelin M, Poole K, Antebi A, Koch M, Stetefeld J
|
| RgGuinier |
5.8 |
nm |
| Dmax |
23.0 |
nm |
| VolumePorod |
142 |
nm3 |
|
|
|
|
|
|
|
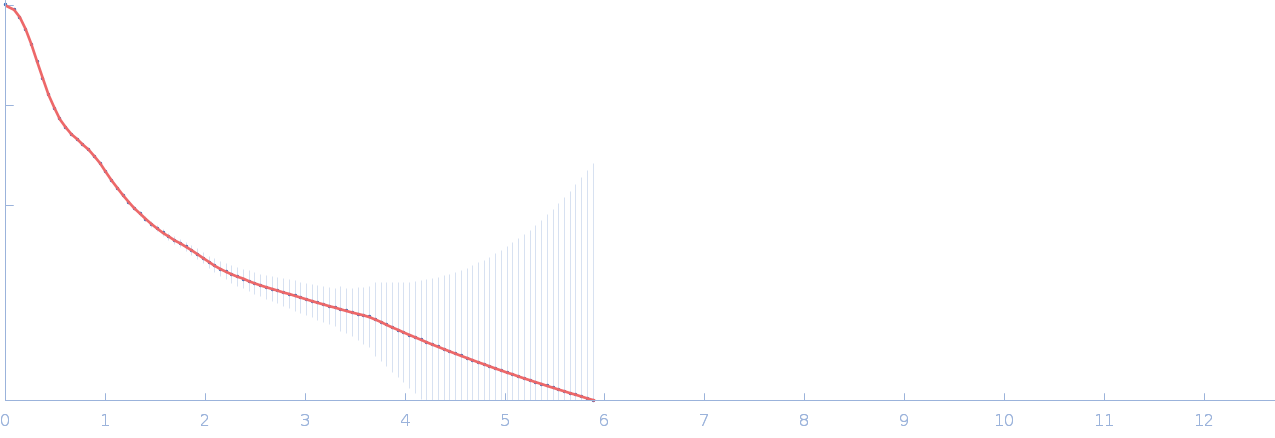
| Sample: |
Netrin-1 pentamer, 248 kDa Gallus gallus protein
Heparin oligosaccharide dp8 ammonium salt monomer, 2 kDa Sus scrofa domesticus
|
| Buffer: |
50 mM Tris-HCl, 200 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Nov 26
|
The dynamic nature of netrin-1 and the structural basis for glycosaminoglycan fragment-induced filament formation
Nature Communications 14(1) (2023)
Meier M, Gupta M, Akgül S, McDougall M, Imhof T, Nikodemus D, Reuten R, Moya-Torres A, To V, Ferens F, Heide F, Padilla-Meier G, Kukura P, Huang W, Gerisch B, Mörgelin M, Poole K, Antebi A, Koch M, Stetefeld J
|
| RgGuinier |
6.5 |
nm |
| Dmax |
23.5 |
nm |
| VolumePorod |
729 |
nm3 |
|
|
|
|
|
|
|
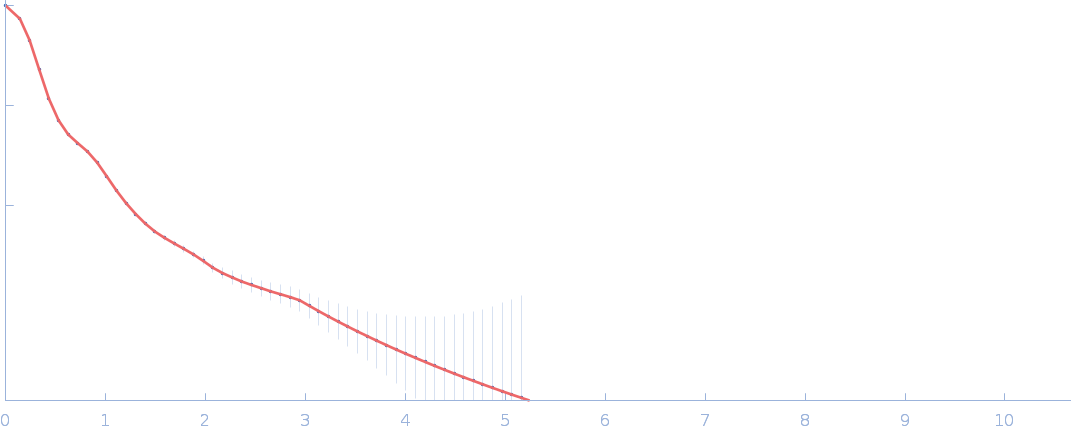
| Sample: |
Netrin-1 pentamer, 248 kDa Gallus gallus protein
Heparin oligosaccharide dp8 ammonium salt monomer, 2 kDa Sus scrofa domesticus
|
| Buffer: |
50 mM Tris-HCl, 200 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Nov 26
|
The dynamic nature of netrin-1 and the structural basis for glycosaminoglycan fragment-induced filament formation
Nature Communications 14(1) (2023)
Meier M, Gupta M, Akgül S, McDougall M, Imhof T, Nikodemus D, Reuten R, Moya-Torres A, To V, Ferens F, Heide F, Padilla-Meier G, Kukura P, Huang W, Gerisch B, Mörgelin M, Poole K, Antebi A, Koch M, Stetefeld J
|
| RgGuinier |
6.4 |
nm |
| Dmax |
21.9 |
nm |
| VolumePorod |
723 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Netrin-1 pentamer, 248 kDa Gallus gallus protein
Heparin oligosaccharide dp10 ammonium salt monomer, 3 kDa Sus scrofa domesticus
|
| Buffer: |
50 mM Tris-HCl, 200 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Nov 26
|
The dynamic nature of netrin-1 and the structural basis for glycosaminoglycan fragment-induced filament formation
Nature Communications 14(1) (2023)
Meier M, Gupta M, Akgül S, McDougall M, Imhof T, Nikodemus D, Reuten R, Moya-Torres A, To V, Ferens F, Heide F, Padilla-Meier G, Kukura P, Huang W, Gerisch B, Mörgelin M, Poole K, Antebi A, Koch M, Stetefeld J
|
| RgGuinier |
6.4 |
nm |
| Dmax |
21.7 |
nm |
| VolumePorod |
758 |
nm3 |
|
|
|
|
|
|
|
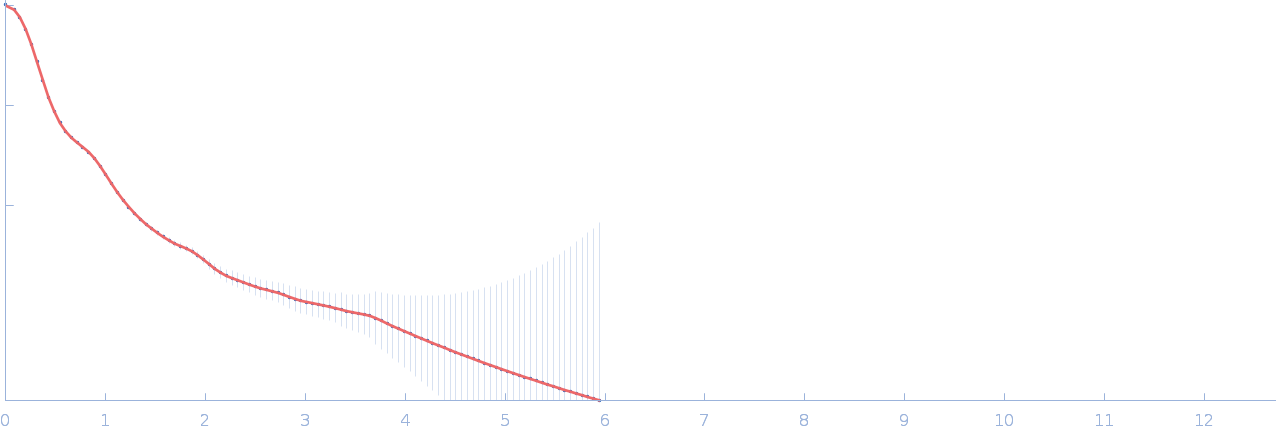
| Sample: |
Netrin-1 pentamer, 248 kDa Gallus gallus protein
Heparin oligosaccharide dp10 ammonium salt monomer, 3 kDa Sus scrofa domesticus
|
| Buffer: |
50 mM Tris-HCl, 200 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Nov 26
|
The dynamic nature of netrin-1 and the structural basis for glycosaminoglycan fragment-induced filament formation
Nature Communications 14(1) (2023)
Meier M, Gupta M, Akgül S, McDougall M, Imhof T, Nikodemus D, Reuten R, Moya-Torres A, To V, Ferens F, Heide F, Padilla-Meier G, Kukura P, Huang W, Gerisch B, Mörgelin M, Poole K, Antebi A, Koch M, Stetefeld J
|
| RgGuinier |
6.5 |
nm |
| Dmax |
21.9 |
nm |
| VolumePorod |
769 |
nm3 |
|
|