|
|
|
|
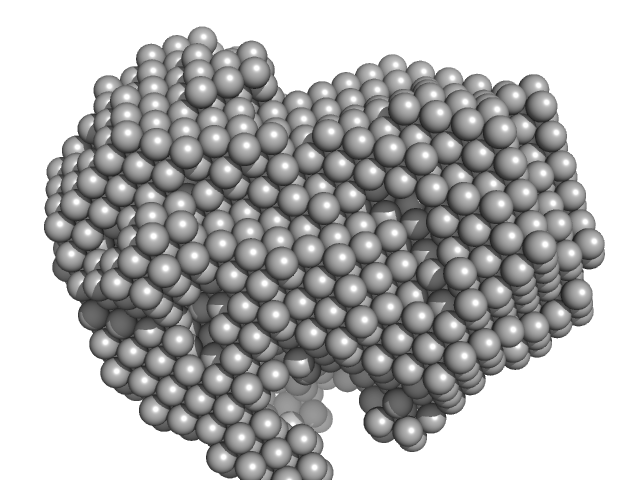
|
| Sample: |
Iron-utilization periplasmic protein monomer, 34 kDa Haemophilus influenzae protein
|
| Buffer: |
10 mM Na2HPO4 . 7H2O 1.8 mM KH2PO4 137 mM NaCl 2.7 mM KCl 5% v/v Glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Aug 20
|
Conformational multiplicity of bacterial ferric binding protein revealed by small angle x-ray scattering and molecular dynamics calculations
The Journal of Chemical Physics 158(8) (2023)
Liu G, Ekmen E, Jalalypour F, Mertens H, Jeffries C, Svergun D, Atilgan A, Atilgan C, Sayers Z
|
| RgGuinier |
2.0 |
nm |
| Dmax |
6.2 |
nm |
| VolumePorod |
34 |
nm3 |
|
|
|
|
|
|
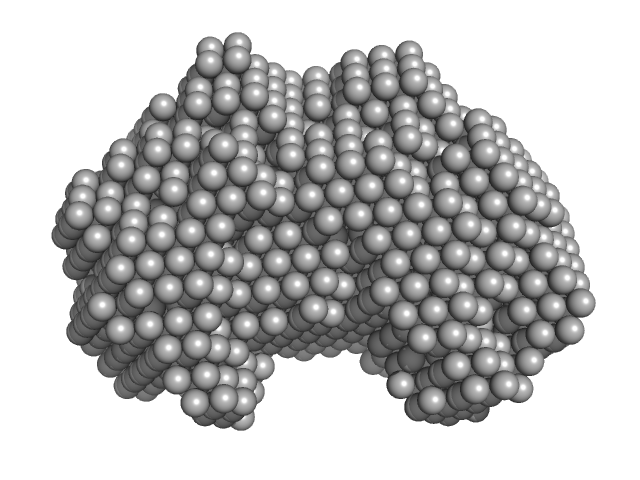
|
| Sample: |
Iron-utilization periplasmic protein monomer, 34 kDa Haemophilus influenzae protein
|
| Buffer: |
10 mM Na2HPO4 . 7H2O 1.8 mM KH2PO4 137 mM NaCl 2.7 mM KCl 5% v/v Glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Dec 9
|
Conformational multiplicity of bacterial ferric binding protein revealed by small angle x-ray scattering and molecular dynamics calculations
The Journal of Chemical Physics 158(8) (2023)
Liu G, Ekmen E, Jalalypour F, Mertens H, Jeffries C, Svergun D, Atilgan A, Atilgan C, Sayers Z
|
| RgGuinier |
2.1 |
nm |
| Dmax |
6.5 |
nm |
| VolumePorod |
39 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Iron-utilization periplasmic protein monomer, 34 kDa Haemophilus influenzae protein
|
| Buffer: |
10 mM Na2HPO4 . 7H2O 1.8 mM KH2PO4 137 mM NaCl 2.7 mM KCl 5% v/v Glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Dec 9
|
Conformational multiplicity of bacterial ferric binding protein revealed by small angle x-ray scattering and molecular dynamics calculations
The Journal of Chemical Physics 158(8) (2023)
Liu G, Ekmen E, Jalalypour F, Mertens H, Jeffries C, Svergun D, Atilgan A, Atilgan C, Sayers Z
|
| RgGuinier |
2.0 |
nm |
| Dmax |
5.9 |
nm |
| VolumePorod |
33 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Iron-utilization periplasmic protein monomer, 34 kDa Haemophilus influenzae protein
|
| Buffer: |
10 mM Na2HPO4 . 7H2O 1.8 mM KH2PO4 137 mM NaCl 2.7 mM KCl 5% v/v Glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Sep 19
|
Conformational multiplicity of bacterial ferric binding protein revealed by small angle x-ray scattering and molecular dynamics calculations
The Journal of Chemical Physics 158(8) (2023)
Liu G, Ekmen E, Jalalypour F, Mertens H, Jeffries C, Svergun D, Atilgan A, Atilgan C, Sayers Z
|
| RgGuinier |
2.1 |
nm |
| Dmax |
6.2 |
nm |
| VolumePorod |
40 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Iron-utilization periplasmic protein monomer, 34 kDa Haemophilus influenzae protein
|
| Buffer: |
1 mM Na2HPO4.7H2O, 0.18 mM KH2PO4, 13.7 mM NaCl, 0.27 mM KCl, 5%v/v Glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Aug 20
|
Conformational multiplicity of bacterial ferric binding protein revealed by small angle x-ray scattering and molecular dynamics calculations
The Journal of Chemical Physics 158(8) (2023)
Liu G, Ekmen E, Jalalypour F, Mertens H, Jeffries C, Svergun D, Atilgan A, Atilgan C, Sayers Z
|
| RgGuinier |
2.0 |
nm |
| Dmax |
6.1 |
nm |
| VolumePorod |
24 |
nm3 |
|
|
|
|
|
|
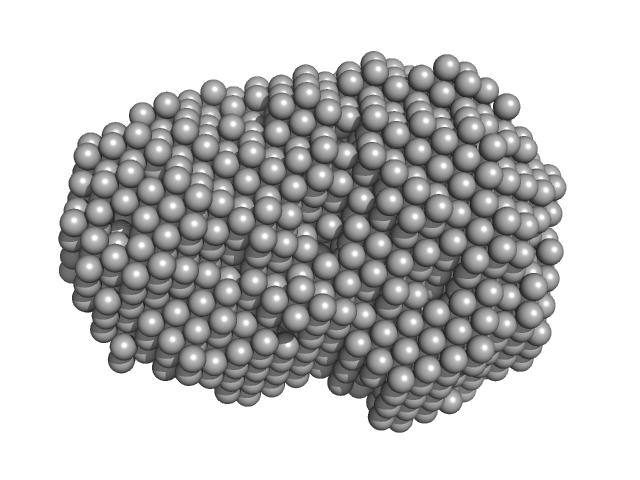
|
| Sample: |
Iron-utilization periplasmic protein monomer, 34 kDa Haemophilus influenzae protein
|
| Buffer: |
1 mM Na2HPO4.7H2O, 0.18 mM KH2PO4, 13.7 mM NaCl, 0.27 mM KCl, 5%v/v Glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Aug 20
|
Conformational multiplicity of bacterial ferric binding protein revealed by small angle x-ray scattering and molecular dynamics calculations
The Journal of Chemical Physics 158(8) (2023)
Liu G, Ekmen E, Jalalypour F, Mertens H, Jeffries C, Svergun D, Atilgan A, Atilgan C, Sayers Z
|
| RgGuinier |
2.1 |
nm |
| Dmax |
6.3 |
nm |
| VolumePorod |
35 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Iron-utilization periplasmic protein monomer, 34 kDa Haemophilus influenzae protein
|
| Buffer: |
1 mM Na2HPO4.7H2O, 0.18 mM KH2PO4, 13.7 mM NaCl, 0.27 mM KCl, 5%v/v Glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Aug 20
|
Conformational multiplicity of bacterial ferric binding protein revealed by small angle x-ray scattering and molecular dynamics calculations
The Journal of Chemical Physics 158(8) (2023)
Liu G, Ekmen E, Jalalypour F, Mertens H, Jeffries C, Svergun D, Atilgan A, Atilgan C, Sayers Z
|
| RgGuinier |
2.0 |
nm |
| Dmax |
6.1 |
nm |
| VolumePorod |
29 |
nm3 |
|
|
|
|
|
|
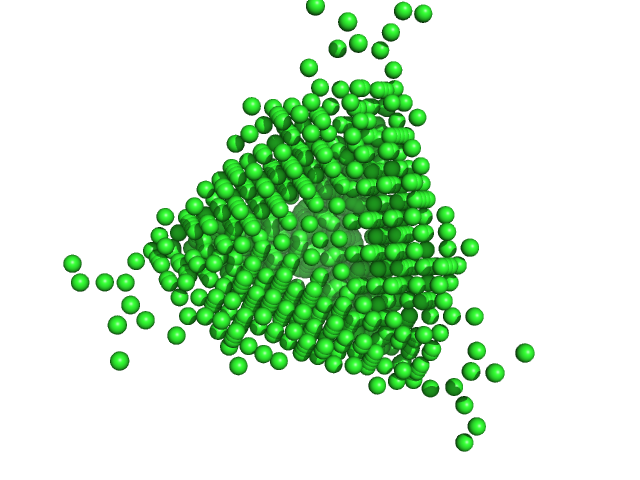
|
| Sample: |
DNA protection during starvation protein dodecamer, 224 kDa Escherichia coli (strain … protein
|
| Buffer: |
50 mM NaCl, 0.5 mM EDTA, 50 mM Tris-HCl, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Oct 5
|
An Anomalous Small-Angle X-Ray Scattering Study of the Formation of Iron Clusters in the Inner Cavity of the Ferritin-Like Protein Dps
Moscow University Physics Bulletin 77(6):858-867 (2023)
Gordienko A, Mozhaev A, Gibizova V, Dadinova L
|
| RgGuinier |
3.8 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
32 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
DNA protection during starvation protein dodecamer, 224 kDa Escherichia coli (strain … protein
|
| Buffer: |
50 mM NaCl, 0.5 mM EDTA, 50 mM Tris-HCl, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Oct 5
|
An Anomalous Small-Angle X-Ray Scattering Study of the Formation of Iron Clusters in the Inner Cavity of the Ferritin-Like Protein Dps
Moscow University Physics Bulletin 77(6):858-867 (2023)
Gordienko A, Mozhaev A, Gibizova V, Dadinova L
|
| RgGuinier |
7.4 |
nm |
| Dmax |
12.0 |
nm |
|
|
|
|
|
|
|
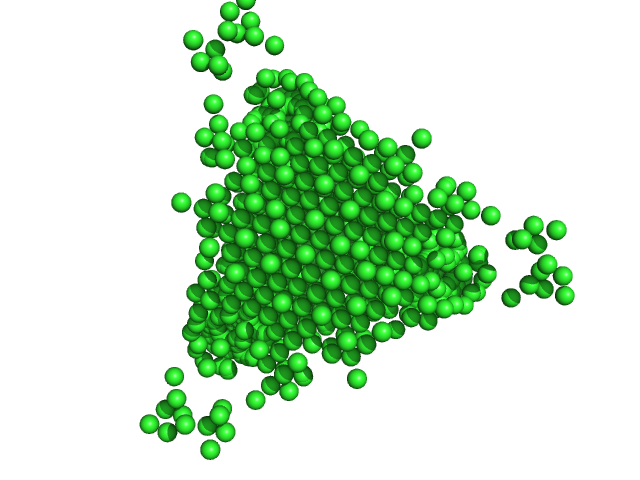
| Sample: |
DNA protection during starvation protein dodecamer, 224 kDa Escherichia coli (strain … protein
|
| Buffer: |
50 mM NaCl, 0.5 mM EDTA, 50 mM Tris-HCl, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Oct 5
|
An Anomalous Small-Angle X-Ray Scattering Study of the Formation of Iron Clusters in the Inner Cavity of the Ferritin-Like Protein Dps
Moscow University Physics Bulletin 77(6):858-867 (2023)
Gordienko A, Mozhaev A, Gibizova V, Dadinova L
|
| RgGuinier |
7.1 |
nm |
| Dmax |
14.0 |
nm |
|
|