|
|
|
|
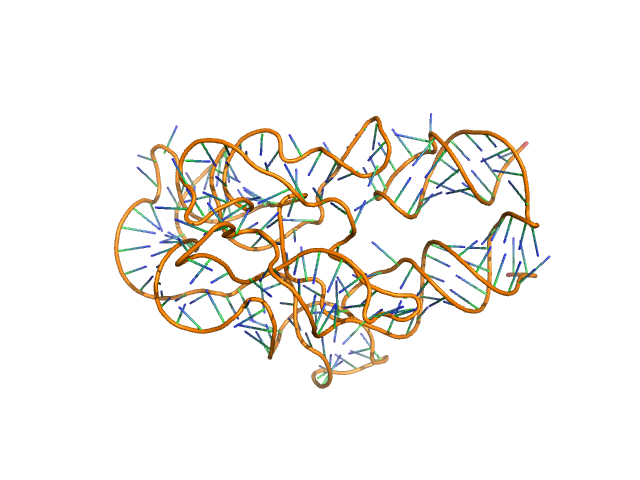
|
| Sample: |
Thermoanearobacter tengcongensis (Tte) fecB riboswitch aptamer domain, 68 kDa marine metagenome RNA
|
| Buffer: |
50 mM MES pH 6.0, 10 mM KCl, 1 mM MgCl2, pH: 6 |
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2021 Apr 18
|
Visualizing RNA conformational and architectural heterogeneity in solution.
Nat Commun 14(1):714 (2023)
Ding J, Lee YT, Bhandari Y, Schwieters CD, Fan L, Yu P, Tarosov SG, Stagno JR, Ma B, Nussinov R, Rein A, Zhang J, Wang YX
|
| RgGuinier |
5.9 |
nm |
| Dmax |
20.7 |
nm |
| VolumePorod |
206 |
nm3 |
|
|
|
|
|
|
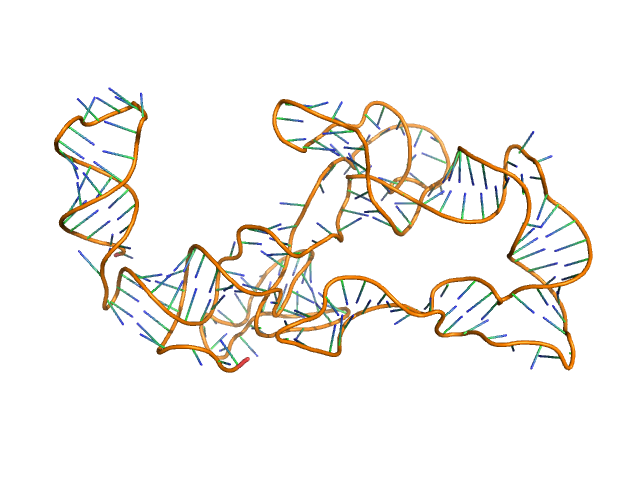
|
| Sample: |
Thermoanearobacter tengcongensis (Tte) fecB riboswitch aptamer domain, 68 kDa marine metagenome RNA
|
| Buffer: |
50 mM MES pH 6.0, 10 mM KCl, 1 mM MgCl2, 4-18 μM coenzyme B12 ligand, pH: 6 |
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2021 Apr 18
|
Visualizing RNA conformational and architectural heterogeneity in solution.
Nat Commun 14(1):714 (2023)
Ding J, Lee YT, Bhandari Y, Schwieters CD, Fan L, Yu P, Tarosov SG, Stagno JR, Ma B, Nussinov R, Rein A, Zhang J, Wang YX
|
| RgGuinier |
5.7 |
nm |
| Dmax |
19.7 |
nm |
| VolumePorod |
230 |
nm3 |
|
|
|
|
|
|

|
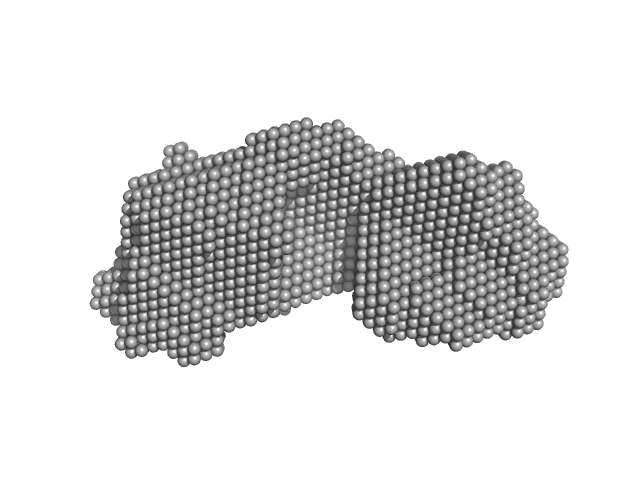
| Sample: |
P123 expressed protein dimer, 248 kDa Mycoplasma mobile (strain … protein
|
| Buffer: |
100 mM ammonium acetate, pH: 6.8 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2019 Oct 25
|
Structure and Function of Gli123 Involved in Mycoplasma mobile Gliding.
J Bacteriol :e0034022 (2023)
Matsuike D, Tahara YO, Nonaka T, Wu HN, Hamaguchi T, Kudo H, Hayashi Y, Arai M, Miyata M
|
| RgGuinier |
8.2 |
nm |
| Dmax |
31.6 |
nm |
| VolumePorod |
436 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
SAVED domain-containing protein monomer, 58 kDa Sulfurihydrogenibium sp. (strain … protein
|
| Buffer: |
20 mM Tris, 50 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Dec 14
|
Antiviral signalling by a cyclic nucleotide activated CRISPR protease.
Nature 614(7946):168-174 (2023)
Rouillon C, Schneberger N, Chi H, Blumenstock K, Da Vela S, Ackermann K, Moecking J, Peter MF, Boenigk W, Seifert R, Bode BE, Schmid-Burgk JL, Svergun D, Geyer M, White MF, Hagelueken G
|
| RgGuinier |
3.1 |
nm |
| Dmax |
10.3 |
nm |
| VolumePorod |
80 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
SAVED domain-containing protein monomer, 58 kDa Sulfurihydrogenibium sp. (strain … protein
|
| Buffer: |
20 mM Tris, 50 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Apr 8
|
Antiviral signalling by a cyclic nucleotide activated CRISPR protease.
Nature 614(7946):168-174 (2023)
Rouillon C, Schneberger N, Chi H, Blumenstock K, Da Vela S, Ackermann K, Moecking J, Peter MF, Boenigk W, Seifert R, Bode BE, Schmid-Burgk JL, Svergun D, Geyer M, White MF, Hagelueken G
|
| RgGuinier |
3.2 |
nm |
| Dmax |
10.3 |
nm |
| VolumePorod |
82 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
SAVED domain-containing protein monomer, 58 kDa Sulfurihydrogenibium sp. (strain … protein
|
| Buffer: |
20 mM Tris, 50 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Apr 8
|
Antiviral signalling by a cyclic nucleotide activated CRISPR protease.
Nature 614(7946):168-174 (2023)
Rouillon C, Schneberger N, Chi H, Blumenstock K, Da Vela S, Ackermann K, Moecking J, Peter MF, Boenigk W, Seifert R, Bode BE, Schmid-Burgk JL, Svergun D, Geyer M, White MF, Hagelueken G
|
| RgGuinier |
3.8 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
116 |
nm3 |
|
|
|
|
|
|

|
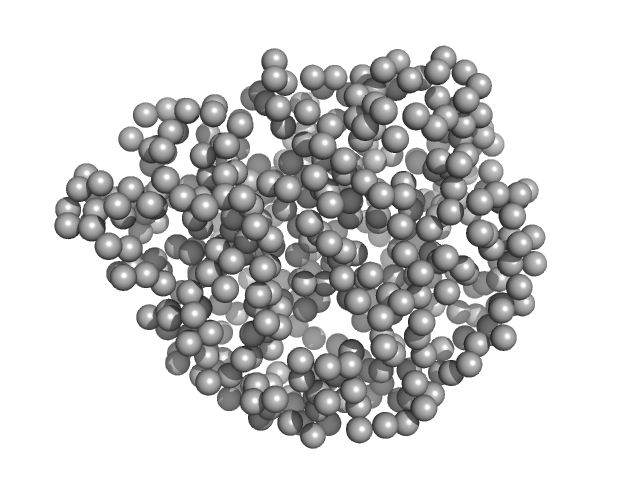
| Sample: |
DNA protection during starvation, DPS (Ferritin superfamily) dodecamer, 270 kDa Deinococcus grandis protein
|
| Buffer: |
50 mM MOPS, 230 mM NaCl, 5 mM EDTA, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Nov 8
|
Controlled modulation of the dynamics of the Deinococcus grandis Dps N-terminal tails by divalent metals.
Protein Sci :e4567 (2023)
Guerra JPL, Blanchet CE, Vieira BJC, Waerenborgh JC, Jones NC, Hoffmann SV, Pereira AS, Tavares P
|
| RgGuinier |
4.5 |
nm |
| Dmax |
19.5 |
nm |
| VolumePorod |
453 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Lanthionine synthetase C-like protein monomer, 50 kDa Clostridium sp. Maddingley … protein
|
| Buffer: |
50 mM HEPES, 500 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at Xenocs Xeuss 2.0 Q-Xoom, Center for Structural Studies, Heinrich-Heine-University on 2019 Nov 28
|
The structure of MadC from Clostridium maddingley reveals new insights into class I lanthipeptide cyclases
Frontiers in Microbiology 13 (2023)
Knospe C, Kamel M, Spitz O, Hoeppner A, Galle S, Reiners J, Kedrov A, Smits S, Schmitt L
|
| RgGuinier |
2.3 |
nm |
| Dmax |
7.8 |
nm |
| VolumePorod |
78 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Lysozyme C monomer, 14 kDa Gallus gallus protein
|
| Buffer: |
100 mM HEPES pH 7.5, 20 %(v/v) jeffamine M-600, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Aug 28
|
The Relationship of Precursor Cluster Concentration in a Saturated Crystallization Solution to Long-Range Order During the Transition to the Solid Phase.
Acta Naturae 15(1):58-68 (2023)
Marchenkova MA, Boikova AS, Ilina KB, Konarev PV, Pisarevsky YV, Dyakova YA, Kovalchuk MV
|
|
|
|
|
|
|
|
| Sample: |
Lysozyme C monomer, 14 kDa Gallus gallus protein
|
| Buffer: |
100 mM sodium acetate, pH 4.6, 2.0 M sodium formate, pH: 4.6 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Aug 28
|
The Relationship of Precursor Cluster Concentration in a Saturated Crystallization Solution to Long-Range Order During the Transition to the Solid Phase.
Acta Naturae 15(1):58-68 (2023)
Marchenkova MA, Boikova AS, Ilina KB, Konarev PV, Pisarevsky YV, Dyakova YA, Kovalchuk MV
|
|
|