|
|
|
|

|
| Sample: |
Diacylglycerol kinase alpha monomer, 22 kDa Homo sapiens protein
|
| Buffer: |
20mM TrisHCl, 200mM NaCl, 5mM DTT, 5% v/v Glycerol, 3mM EGTA, pH: 7.4 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2020 Dec 8
|
Ca 2+
‐induced structural changes and intramolecular interactions in N‐terminal region of diacylglycerol kinase alpha
Protein Science 31(7) (2022)
Takahashi D, Yonezawa K, Okizaki Y, Caaveiro J, Ueda T, Shimada A, Sakane F, Shimizu N
|
| RgGuinier |
1.9 |
nm |
| Dmax |
7.0 |
nm |
| VolumePorod |
44 |
nm3 |
|
|
|
|
|
|

|
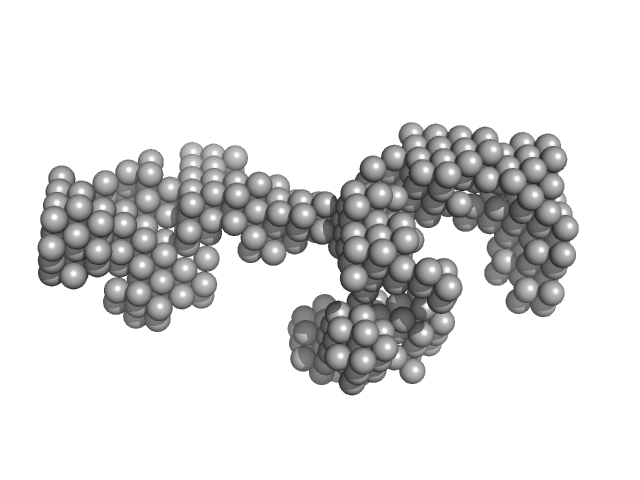
| Sample: |
LincRNA-p21 AluSx1 Sense RNA monomer, 100 kDa Homo sapiens RNA
|
| Buffer: |
50mM HEPES,150 mM NaCl, 15 mM MgCl2, 3% glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2021 Jul 1
|
Biophysical characterisation of human LincRNA-p21 sense and antisense Alu inverted repeats.
Nucleic Acids Res 50(10):5881-5898 (2022)
D'Souza MH, Mrozowich T, Badmalia MD, Geeraert M, Frederickson A, Henrickson A, Demeler B, Wolfinger MT, Patel TR
|
| RgGuinier |
6.1 |
nm |
| Dmax |
18.5 |
nm |
| VolumePorod |
225 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
LincRNA-p21 AluSx1 Antisense RNA monomer, 90 kDa Homo sapiens RNA
|
| Buffer: |
50mM HEPES,150 mM NaCl, 15 mM MgCl2, 3% glycerol, pH: 7.4 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2021 Jul 1
|
Biophysical characterisation of human LincRNA-p21 sense and antisense Alu inverted repeats.
Nucleic Acids Res 50(10):5881-5898 (2022)
D'Souza MH, Mrozowich T, Badmalia MD, Geeraert M, Frederickson A, Henrickson A, Demeler B, Wolfinger MT, Patel TR
|
| RgGuinier |
5.9 |
nm |
| Dmax |
18.1 |
nm |
| VolumePorod |
375 |
nm3 |
|
|
|
|
|
|
|
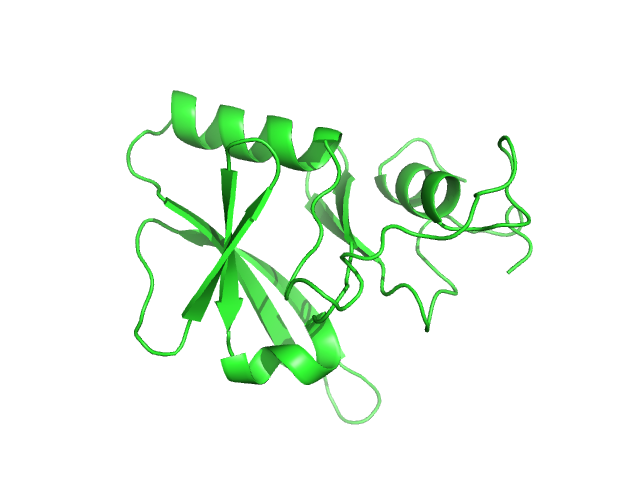
| Sample: |
Iron-sulfur cluster assembly 1 homolog, mitochondrial, 15 kDa Columba livia protein
|
| Buffer: |
20 mM Tris-HCl, 0.15 M NaCl, 10 mM 3-mercapto-1,2-propanediol, pH: 8 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2020 Feb 25
|
Magnetic field effects on the structure and molecular behavior of pigeon iron–sulfur protein
Protein Science 31(6) (2022)
Arai S, Shimizu R, Adachi M, Hirai M
|
|
|
|
|
|
|
|
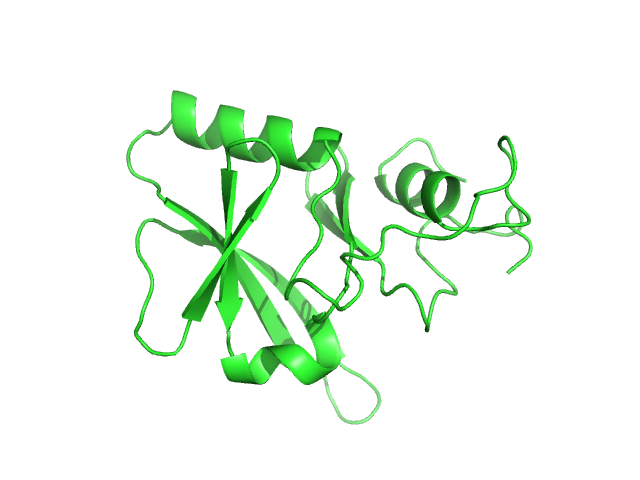
| Sample: |
Iron-sulfur cluster assembly 1 homolog, mitochondrial, 15 kDa Columba livia protein
|
| Buffer: |
20 mM Tris-HCl, 0.15 M NaCl, 10 mM 3-mercapto-1,2-propanediol, pH: 8 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2020 Feb 25
|
Magnetic field effects on the structure and molecular behavior of pigeon iron–sulfur protein
Protein Science 31(6) (2022)
Arai S, Shimizu R, Adachi M, Hirai M
|
|
|
|
|
|
|
|
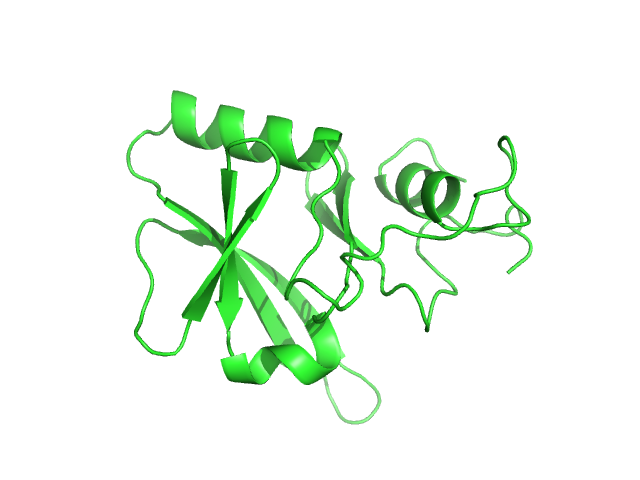
| Sample: |
Iron-sulfur cluster assembly 1 homolog, mitochondrial, 15 kDa Columba livia protein
|
| Buffer: |
20 mM Tris-HCl, 0.15 M NaCl, 10 mM 3-mercapto-1,2-propanediol, pH: 8 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2020 Feb 25
|
Magnetic field effects on the structure and molecular behavior of pigeon iron–sulfur protein
Protein Science 31(6) (2022)
Arai S, Shimizu R, Adachi M, Hirai M
|
|
|
|
|
|
|
|
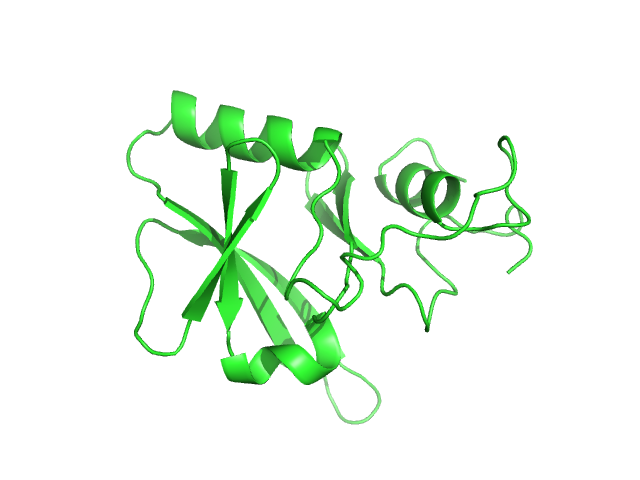
| Sample: |
Iron-sulfur cluster assembly 1 homolog, mitochondrial, 15 kDa Columba livia protein
|
| Buffer: |
20 mM Tris-HCl, 0.15 M NaCl, 10 mM 3-mercapto-1,2-propanediol, pH: 8 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2020 Feb 25
|
Magnetic field effects on the structure and molecular behavior of pigeon iron–sulfur protein
Protein Science 31(6) (2022)
Arai S, Shimizu R, Adachi M, Hirai M
|
|
|
|
|
|
|
|
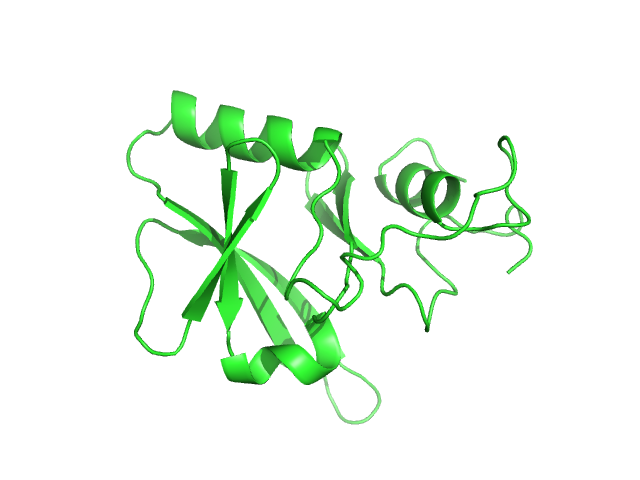
| Sample: |
Iron-sulfur cluster assembly 1 homolog, mitochondrial, 15 kDa Columba livia protein
|
| Buffer: |
20 mM Tris-HCl, 0.15 M NaCl, 10 mM 3-mercapto-1,2-propanediol, pH: 8 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2020 Feb 25
|
Magnetic field effects on the structure and molecular behavior of pigeon iron–sulfur protein
Protein Science 31(6) (2022)
Arai S, Shimizu R, Adachi M, Hirai M
|
|
|
|
|
|
|
|
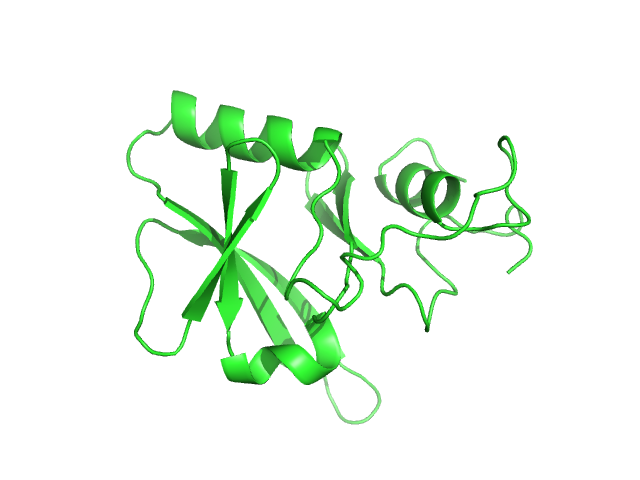
| Sample: |
Iron-sulfur cluster assembly 1 homolog, mitochondrial, 15 kDa Columba livia protein
|
| Buffer: |
20 mM Tris-HCl, 0.15 M NaCl, 10 mM 3-mercapto-1,2-propanediol, pH: 8 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2020 Feb 25
|
Magnetic field effects on the structure and molecular behavior of pigeon iron–sulfur protein
Protein Science 31(6) (2022)
Arai S, Shimizu R, Adachi M, Hirai M
|
|
|
|
|
|
|
|
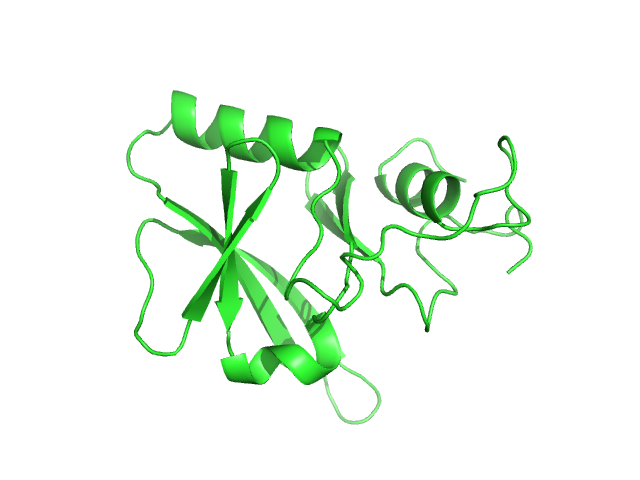
| Sample: |
Iron-sulfur cluster assembly 1 homolog, mitochondrial, 15 kDa Columba livia protein
|
| Buffer: |
20 mM Tris-HCl, 0.15 M NaCl, 10 mM 3-mercapto-1,2-propanediol, pH: 8 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2020 Feb 25
|
Magnetic field effects on the structure and molecular behavior of pigeon iron–sulfur protein
Protein Science 31(6) (2022)
Arai S, Shimizu R, Adachi M, Hirai M
|
|
|