|
|
|
|

|
| Sample: |
Polyketide synthase Pks13 monomer, 33 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
50 mM Tris-HCl, 50 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2006 Nov 1
|
Solution structure of the type I polyketide synthase Pks13 from Mycobacterium tuberculosis.
BMC Biol 20(1):147 (2022)
Bon C, Cabantous S, Julien S, Guillet V, Chalut C, Rima J, Brison Y, Malaga W, Sanchez-Dafun A, Gavalda S, Quémard A, Marcoux J, Waldo GS, Guilhot C, Mourey L
|
| RgGuinier |
1.9 |
nm |
| Dmax |
5.5 |
nm |
| VolumePorod |
56 |
nm3 |
|
|
|
|
|
|
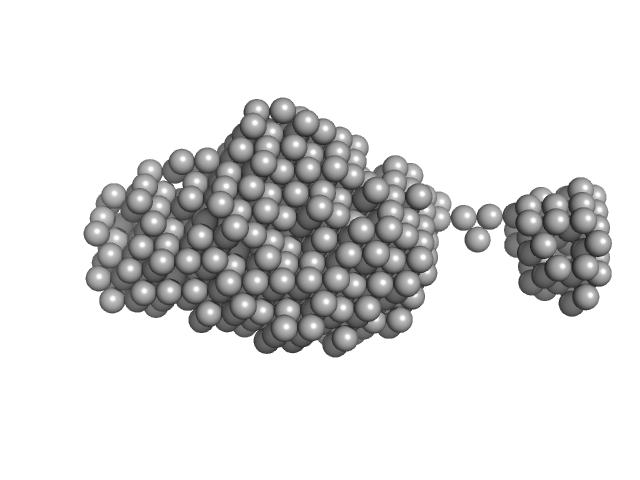
|
| Sample: |
Polyketide synthase Pks13 monomer, 62 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
50 mM Tris-HCl 50 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2006 Nov 1
|
Solution structure of the type I polyketide synthase Pks13 from Mycobacterium tuberculosis.
BMC Biol 20(1):147 (2022)
Bon C, Cabantous S, Julien S, Guillet V, Chalut C, Rima J, Brison Y, Malaga W, Sanchez-Dafun A, Gavalda S, Quémard A, Marcoux J, Waldo GS, Guilhot C, Mourey L
|
| RgGuinier |
3.2 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
80 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Polyketide synthase Pks13 monomer, 53 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
50 mM Tris-HCl, 300 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2006 Nov 1
|
Solution structure of the type I polyketide synthase Pks13 from Mycobacterium tuberculosis.
BMC Biol 20(1):147 (2022)
Bon C, Cabantous S, Julien S, Guillet V, Chalut C, Rima J, Brison Y, Malaga W, Sanchez-Dafun A, Gavalda S, Quémard A, Marcoux J, Waldo GS, Guilhot C, Mourey L
|
| RgGuinier |
3.5 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
115 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Polyketide synthase Pks13 monomer, 51 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
50 mM Tris-HCl, 300 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2011 Jun 3
|
Solution structure of the type I polyketide synthase Pks13 from Mycobacterium tuberculosis.
BMC Biol 20(1):147 (2022)
Bon C, Cabantous S, Julien S, Guillet V, Chalut C, Rima J, Brison Y, Malaga W, Sanchez-Dafun A, Gavalda S, Quémard A, Marcoux J, Waldo GS, Guilhot C, Mourey L
|
| RgGuinier |
2.8 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
85 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Polyketide synthase Pks13 monomer, 116 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
50 mM Tris-HCl 50 mM NaCl, pH: 8.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2006 Nov 1
|
Solution structure of the type I polyketide synthase Pks13 from Mycobacterium tuberculosis.
BMC Biol 20(1):147 (2022)
Bon C, Cabantous S, Julien S, Guillet V, Chalut C, Rima J, Brison Y, Malaga W, Sanchez-Dafun A, Gavalda S, Quémard A, Marcoux J, Waldo GS, Guilhot C, Mourey L
|
| RgGuinier |
3.8 |
nm |
| Dmax |
12.9 |
nm |
| VolumePorod |
186 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Polyketide synthase Pks13 monomer, 51 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
50 mM Tris-HCl, 50 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2012 Dec 7
|
Solution structure of the type I polyketide synthase Pks13 from Mycobacterium tuberculosis.
BMC Biol 20(1):147 (2022)
Bon C, Cabantous S, Julien S, Guillet V, Chalut C, Rima J, Brison Y, Malaga W, Sanchez-Dafun A, Gavalda S, Quémard A, Marcoux J, Waldo GS, Guilhot C, Mourey L
|
| RgGuinier |
2.9 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
82 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Polyketide synthase Pks13 monomer, 104 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
50 mM Tris-HCl, 50 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2008 Sep 20
|
Solution structure of the type I polyketide synthase Pks13 from Mycobacterium tuberculosis.
BMC Biol 20(1):147 (2022)
Bon C, Cabantous S, Julien S, Guillet V, Chalut C, Rima J, Brison Y, Malaga W, Sanchez-Dafun A, Gavalda S, Quémard A, Marcoux J, Waldo GS, Guilhot C, Mourey L
|
| RgGuinier |
3.8 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
155 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Mycocerosic acid synthase monomer, 224 kDa Mycobacterium bovis (strain … protein
|
| Buffer: |
50 mM Tris-HCl, 50 mM NaCl, 10% glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2013 Oct 1
|
Solution structure of the type I polyketide synthase Pks13 from Mycobacterium tuberculosis.
BMC Biol 20(1):147 (2022)
Bon C, Cabantous S, Julien S, Guillet V, Chalut C, Rima J, Brison Y, Malaga W, Sanchez-Dafun A, Gavalda S, Quémard A, Marcoux J, Waldo GS, Guilhot C, Mourey L
|
| RgGuinier |
5.7 |
nm |
| Dmax |
21.0 |
nm |
| VolumePorod |
420 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Phenolpthiocerol synthesis type-I polyketide synthase ppsA monomer, 199 kDa Mycobacterium bovis (strain … protein
|
| Buffer: |
50 mM Tris-HCl, 500 mM NaCl, 2 mM EDTA, 10% glycerol, pH: 8 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2012 Jun 14
|
Solution structure of the type I polyketide synthase Pks13 from Mycobacterium tuberculosis.
BMC Biol 20(1):147 (2022)
Bon C, Cabantous S, Julien S, Guillet V, Chalut C, Rima J, Brison Y, Malaga W, Sanchez-Dafun A, Gavalda S, Quémard A, Marcoux J, Waldo GS, Guilhot C, Mourey L
|
| RgGuinier |
6.7 |
nm |
| Dmax |
20.2 |
nm |
| VolumePorod |
680 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Diacylglycerol kinase alpha monomer, 22 kDa Homo sapiens protein
|
| Buffer: |
20 mM TrisHCl, 200 mM NaCl, 5 mM DTT, 5% v/v glycerol, 3 mM CaCl2, pH: 7.4 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2020 Dec 8
|
Ca 2+
‐induced structural changes and intramolecular interactions in N‐terminal region of diacylglycerol kinase alpha
Protein Science 31(7) (2022)
Takahashi D, Yonezawa K, Okizaki Y, Caaveiro J, Ueda T, Shimada A, Sakane F, Shimizu N
|
| RgGuinier |
1.9 |
nm |
| Dmax |
6.2 |
nm |
| VolumePorod |
44 |
nm3 |
|
|