|
|
|
|

|
| Sample: |
Ganglioside-induced differentiation-associated protein 1 (H123R) dimer, 70 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 300 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2020 Oct 8
|
Structural insights into Charcot-Marie-Tooth disease-linked mutations in human GDAP1.
FEBS Open Bio (2022)
Sutinen A, Nguyen GTT, Raasakka A, Muruganandam G, Loris R, Ylikallio E, Tyynismaa H, Bartesaghi L, Ruskamo S, Kursula P
|
| RgGuinier |
3.1 |
nm |
| Dmax |
9.9 |
nm |
| VolumePorod |
107 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Ganglioside-induced differentiation-associated protein 1 (R120W) dimer, 70 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 300 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2020 Jul 1
|
Structural insights into Charcot-Marie-Tooth disease-linked mutations in human GDAP1.
FEBS Open Bio (2022)
Sutinen A, Nguyen GTT, Raasakka A, Muruganandam G, Loris R, Ylikallio E, Tyynismaa H, Bartesaghi L, Ruskamo S, Kursula P
|
| RgGuinier |
3.1 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
105 |
nm3 |
|
|
|
|
|
|

|
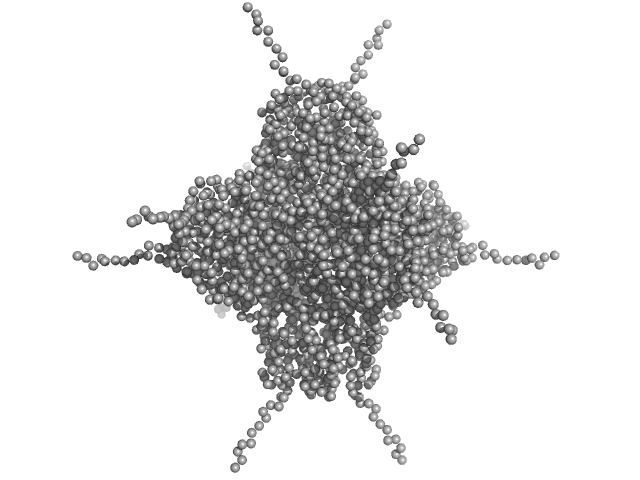
| Sample: |
DNA protection during starvation, DPS (Ferritin superfamily) dodecamer, 270 kDa Deinococcus grandis protein
|
| Buffer: |
50 mM MOPS-NaOH, 50 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Oct 22
|
The Conformation of the N-Terminal Tails of Deinococcus grandis Dps Is Modulated by the Ionic Strength
International Journal of Molecular Sciences 23(9):4871 (2022)
Guerra J, Blanchet C, Vieira B, Almeida A, Waerenborgh J, Jones N, Hoffmann S, Tavares P, Pereira A
|
| RgGuinier |
4.4 |
nm |
| Dmax |
16.1 |
nm |
| VolumePorod |
409 |
nm3 |
|
|
|
|
|
|
|
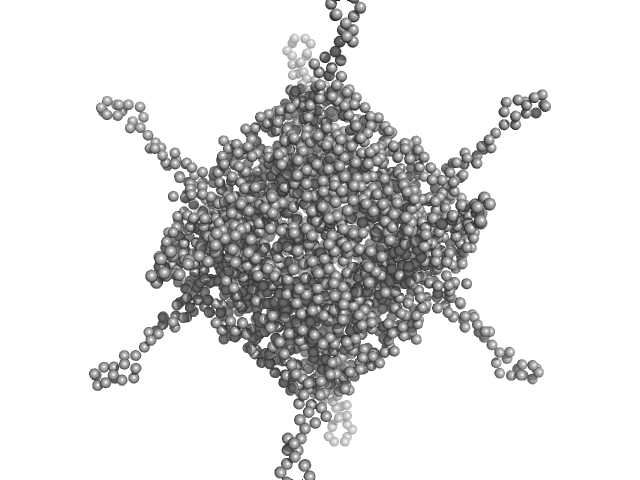
| Sample: |
DNA protection during starvation, DPS (Ferritin superfamily) dodecamer, 270 kDa Deinococcus grandis protein
|
| Buffer: |
50 mM MOPS-NaOH, 80 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Oct 22
|
The Conformation of the N-Terminal Tails of Deinococcus grandis Dps Is Modulated by the Ionic Strength
International Journal of Molecular Sciences 23(9):4871 (2022)
Guerra J, Blanchet C, Vieira B, Almeida A, Waerenborgh J, Jones N, Hoffmann S, Tavares P, Pereira A
|
| RgGuinier |
4.5 |
nm |
| Dmax |
17.1 |
nm |
| VolumePorod |
475 |
nm3 |
|
|
|
|
|
|
|
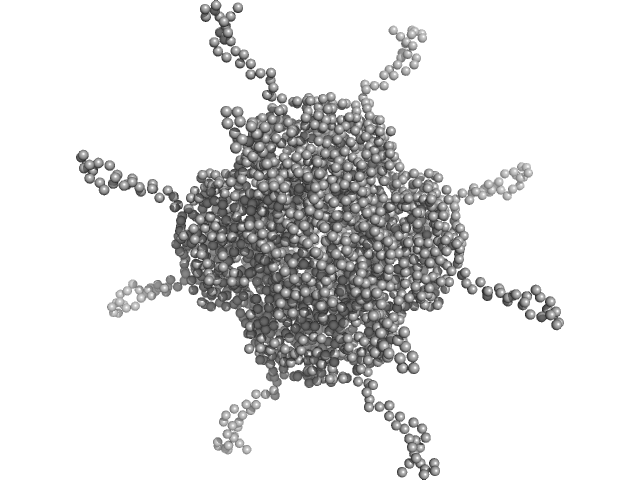
| Sample: |
DNA protection during starvation, DPS (Ferritin superfamily) dodecamer, 270 kDa Deinococcus grandis protein
|
| Buffer: |
50 mM MOPS-NaOH, 230 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Oct 22
|
The Conformation of the N-Terminal Tails of Deinococcus grandis Dps Is Modulated by the Ionic Strength
International Journal of Molecular Sciences 23(9):4871 (2022)
Guerra J, Blanchet C, Vieira B, Almeida A, Waerenborgh J, Jones N, Hoffmann S, Tavares P, Pereira A
|
| RgGuinier |
4.5 |
nm |
| Dmax |
20.6 |
nm |
| VolumePorod |
438 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
DNA protection during starvation, DPS (Ferritin superfamily) dodecamer, 270 kDa Deinococcus grandis protein
|
| Buffer: |
50 mM MOPS-NaOH, 480 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Oct 22
|
The Conformation of the N-Terminal Tails of Deinococcus grandis Dps Is Modulated by the Ionic Strength
International Journal of Molecular Sciences 23(9):4871 (2022)
Guerra J, Blanchet C, Vieira B, Almeida A, Waerenborgh J, Jones N, Hoffmann S, Tavares P, Pereira A
|
| RgGuinier |
4.8 |
nm |
| Dmax |
20.5 |
nm |
| VolumePorod |
430 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
DNA protection during starvation, DPS-∆N (Ferritin superfamily) dodecamer, 218 kDa Deinococcus grandis protein
|
| Buffer: |
50 mM MOPS-NaOH, 50 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Oct 22
|
The Conformation of the N-Terminal Tails of Deinococcus grandis Dps Is Modulated by the Ionic Strength
International Journal of Molecular Sciences 23(9):4871 (2022)
Guerra J, Blanchet C, Vieira B, Almeida A, Waerenborgh J, Jones N, Hoffmann S, Tavares P, Pereira A
|
| RgGuinier |
3.8 |
nm |
| Dmax |
9.3 |
nm |
| VolumePorod |
290 |
nm3 |
|
|
|
|
|
|

|
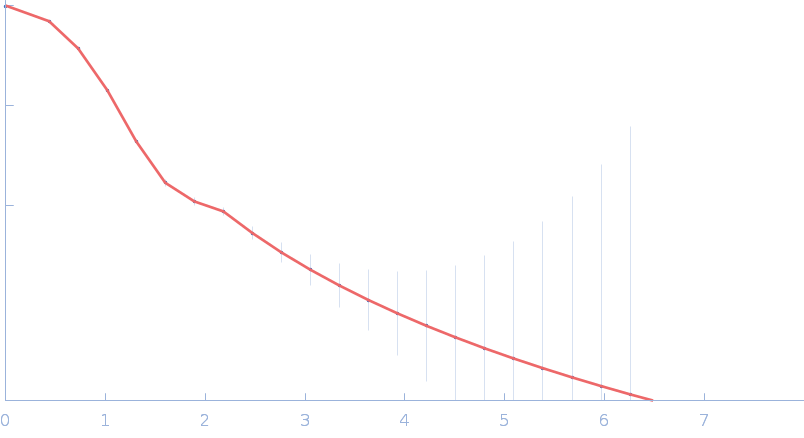
| Sample: |
DNA protection during starvation, DPS-∆N (Ferritin superfamily) dodecamer, 218 kDa Deinococcus grandis protein
|
| Buffer: |
50 mM MOPS-NaOH, 230 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Oct 22
|
The Conformation of the N-Terminal Tails of Deinococcus grandis Dps Is Modulated by the Ionic Strength
International Journal of Molecular Sciences 23(9):4871 (2022)
Guerra J, Blanchet C, Vieira B, Almeida A, Waerenborgh J, Jones N, Hoffmann S, Tavares P, Pereira A
|
| RgGuinier |
3.8 |
nm |
| Dmax |
8.8 |
nm |
| VolumePorod |
291 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Sulfite reductase [NADPH] hemoprotein beta-component (Assimilatory NADPH-dependent sulfite reductase hemoprotein) monomer, 64 kDa Escherichia coli (strain … protein
|
| Buffer: |
50 mM KPi, 100 mM NaCl, 1 mM EDTA, pH: 7.8 |
| Experiment: |
SANS
data collected at EQ-SANS (BL-6), Spallation Neutron Source on 2018 Jul 13
|
Neutron scattering maps the higher-order assembly of NADPH-dependent assimilatory sulfite reductase.
Biophys J (2022)
Murray DT, Walia N, Weiss KL, Stanley CB, Randolph PS, Nagy G, Stroupe ME
|
| RgGuinier |
2.3 |
nm |
| Dmax |
7.2 |
nm |
| VolumePorod |
71 |
nm3 |
|
|
|
|
|
|
|
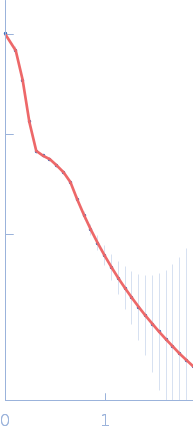
| Sample: |
Sulfite reductase [NADPH] hemoprotein beta-component tetramer, 256 kDa Escherichia coli (strain … protein
Sulfite reductase [NADPH] flavoprotein alpha-component (His-tagged) octamer, 565 kDa Escherichia coli (strain … protein
|
| Buffer: |
50 mM KPi, 100 mM NaCl, 1 mM EDTA, pH: 7.8 |
| Experiment: |
SANS
data collected at EQ-SANS (BL-6), Spallation Neutron Source on 2021 Apr 3
|
Neutron scattering maps the higher-order assembly of NADPH-dependent assimilatory sulfite reductase.
Biophys J (2022)
Murray DT, Walia N, Weiss KL, Stanley CB, Randolph PS, Nagy G, Stroupe ME
|
| RgGuinier |
11.2 |
nm |
| Dmax |
30.5 |
nm |
|
|