|
|
|
|
|
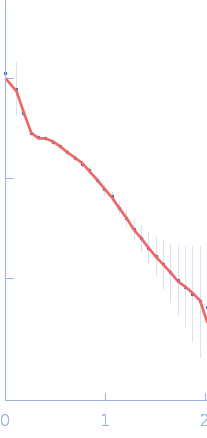
| Sample: |
Sulfite reductase [NADPH] hemoprotein beta-component tetramer, 256 kDa Escherichia coli (strain … protein
Sulfite reductase [NADPH] flavoprotein alpha-component (His-tagged) octamer, 565 kDa Escherichia coli (strain … protein
|
| Buffer: |
50 mM KPi, 100 mM NaCl, 1 mM EDTA, pH: 7.8 |
| Experiment: |
SANS
data collected at EQ-SANS (BL-6), Spallation Neutron Source on 2021 Apr 3
|
Neutron scattering maps the higher-order assembly of NADPH-dependent assimilatory sulfite reductase.
Biophys J (2022)
Murray DT, Walia N, Weiss KL, Stanley CB, Randolph PS, Nagy G, Stroupe ME
|
| RgGuinier |
11.4 |
nm |
| Dmax |
28.5 |
nm |
| VolumePorod |
417 |
nm3 |
|
|
|
|
|
|
|
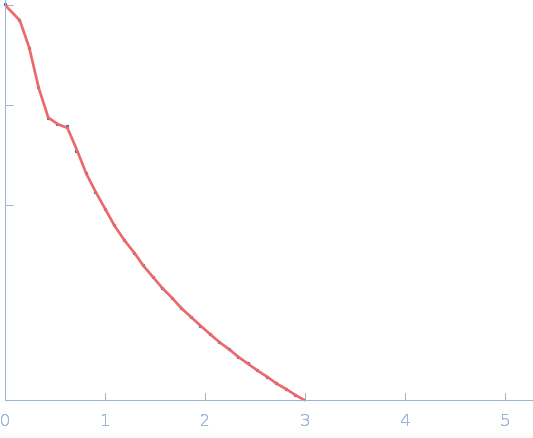
| Sample: |
Sulfite reductase [NADPH] hemoprotein beta-component tetramer, 256 kDa Escherichia coli (strain … protein
Sulfite reductase [NADPH] flavoprotein alpha-component (His-tagged) octamer, 565 kDa Escherichia coli (strain … protein
|
| Buffer: |
50 mM KPi, 100 mM NaCl, 1 mM EDTA, pH: 7.8 |
| Experiment: |
SANS
data collected at EQ-SANS (BL-6), Spallation Neutron Source on 2021 Apr 3
|
Neutron scattering maps the higher-order assembly of NADPH-dependent assimilatory sulfite reductase.
Biophys J (2022)
Murray DT, Walia N, Weiss KL, Stanley CB, Randolph PS, Nagy G, Stroupe ME
|
| RgGuinier |
6.8 |
nm |
| Dmax |
22.0 |
nm |
| VolumePorod |
460 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Type I restriction-modification system methyltransferase subunit dimer, 144 kDa Vibrio vulnificus (strain … protein
Protein Ocr dimer, 28 kDa Escherichia phage T7 protein
|
| Buffer: |
20 mM Tris-HCl,150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at 4C, Pohang Accelerator Laboratory on 2020 Apr 22
|
Structural features of a minimal intact methyltransferase of a type I restriction-modification system.
Int J Biol Macromol (2022)
Seo PW, Hofmann A, Kim JH, Hwangbo SA, Kim JH, Kim JW, Huynh TYL, Choy HE, Kim SJ, Lee J, Lee JO, Jin KS, Park SY, Kim JS
|
| RgGuinier |
4.2 |
nm |
| Dmax |
15.9 |
nm |
| VolumePorod |
354 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Circadian clock protein KaiA dodecamer, 393 kDa Synechococcus elongatus (strain … protein
Circadian clock protein KaiB hexamer, 71 kDa Synechococcus elongatus (strain … protein
Circadian clock protein kinase KaiC (S431D mutant) hexamer, 357 kDa Synechococcus elongatus (strain … protein
|
| Buffer: |
50 mM sodium phosphate buffer, 150 mM NaCl, 5 mM MgCl2, 0.5 mM EDTA, 1 mM ATP, 1 mM DTT, 50 mM arginine, 50 mM glutamine, pH: 7.8 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2017 Dec 8
|
Overall structure of fully assembled cyanobacterial KaiABC circadian clock complex by an integrated experimental-computational approach.
Commun Biol 5(1):184 (2022)
Yunoki Y, Matsumoto A, Morishima K, Martel A, Porcar L, Sato N, Yogo R, Tominaga T, Inoue R, Yagi-Utsumi M, Okuda A, Shimizu M, Urade R, Terauchi K, Kono H, Yagi H, Kato K, Sugiyama M
|
| RgGuinier |
7.0 |
nm |
| Dmax |
24.5 |
nm |
| VolumePorod |
1650 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
75% deuterated Circadian clock protein KaiB hexamer, 71 kDa Synechococcus elongatus (strain … protein
Circadian clock protein KaiA dodecamer, 393 kDa Synechococcus elongatus (strain … protein
75% deuterated Circadian clock protein kinase KaiC (S431D mutant) hexamer, 357 kDa Synechococcus elongatus (strain … protein
|
| Buffer: |
50 mM sodium phosphate buffer, 150 mm NaCl, 5 mM MgCl2, 0.5 mM EDTA, 1 mM ATP, 1 mM DTT, 50 mM arginine, 50 mM glutamine, in 100% D2O, pH: 7.8 |
| Experiment: |
SANS
data collected at D22, Institut Laue-Langevin (ILL) on 2018 Sep 19
|
Overall structure of fully assembled cyanobacterial KaiABC circadian clock complex by an integrated experimental-computational approach.
Commun Biol 5(1):184 (2022)
Yunoki Y, Matsumoto A, Morishima K, Martel A, Porcar L, Sato N, Yogo R, Tominaga T, Inoue R, Yagi-Utsumi M, Okuda A, Shimizu M, Urade R, Terauchi K, Kono H, Yagi H, Kato K, Sugiyama M
|
| RgGuinier |
7.8 |
nm |
| Dmax |
25.6 |
nm |
| VolumePorod |
1620 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Accessory colonization factor monomer, 160 kDa Escherichia coli (strain … protein
|
| Buffer: |
20 mM citrate-phosphate buffer, 200 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Jul 27
|
Molecular and cellular insight into Escherichia coli SslE and its role during biofilm maturation
npj Biofilms and Microbiomes 8(1) (2022)
Corsini P, Wang S, Rehman S, Fenn K, Sagar A, Sirovica S, Cleaver L, Edwards-Gayle C, Mastroianni G, Dorgan B, Sewell L, Lynham S, Iuga D, Franks W, Jarvis J, Carpenter G, Curtis M, Bernadó P, Darbari V, Garnett J
|
| RgGuinier |
4.0 |
nm |
| Dmax |
14.1 |
nm |
| VolumePorod |
244 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Accessory colonization factor monomer, 160 kDa Escherichia coli (strain … protein
|
| Buffer: |
20 mM citrate-phosphate buffer, 200 mM NaCl, pH: 4.4 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Jul 27
|
Molecular and cellular insight into Escherichia coli SslE and its role during biofilm maturation
npj Biofilms and Microbiomes 8(1) (2022)
Corsini P, Wang S, Rehman S, Fenn K, Sagar A, Sirovica S, Cleaver L, Edwards-Gayle C, Mastroianni G, Dorgan B, Sewell L, Lynham S, Iuga D, Franks W, Jarvis J, Carpenter G, Curtis M, Bernadó P, Darbari V, Garnett J
|
| RgGuinier |
3.9 |
nm |
| Dmax |
13.7 |
nm |
| VolumePorod |
248 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Accessory colonization factor monomer, 17 kDa Escherichia coli (strain … protein
|
| Buffer: |
20 mM Tris, 200 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2021 Apr 21
|
Molecular and cellular insight into Escherichia coli SslE and its role during biofilm maturation
npj Biofilms and Microbiomes 8(1) (2022)
Corsini P, Wang S, Rehman S, Fenn K, Sagar A, Sirovica S, Cleaver L, Edwards-Gayle C, Mastroianni G, Dorgan B, Sewell L, Lynham S, Iuga D, Franks W, Jarvis J, Carpenter G, Curtis M, Bernadó P, Darbari V, Garnett J
|
| RgGuinier |
2.0 |
nm |
| Dmax |
7.0 |
nm |
| VolumePorod |
28 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Accessory colonization factor monomer, 23 kDa Escherichia coli (strain … protein
|
| Buffer: |
20 mM Tris, 200 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2021 Apr 21
|
Molecular and cellular insight into Escherichia coli SslE and its role during biofilm maturation
npj Biofilms and Microbiomes 8(1) (2022)
Corsini P, Wang S, Rehman S, Fenn K, Sagar A, Sirovica S, Cleaver L, Edwards-Gayle C, Mastroianni G, Dorgan B, Sewell L, Lynham S, Iuga D, Franks W, Jarvis J, Carpenter G, Curtis M, Bernadó P, Darbari V, Garnett J
|
| RgGuinier |
2.2 |
nm |
| Dmax |
7.7 |
nm |
| VolumePorod |
42 |
nm3 |
|
|
|
|
|
|
|
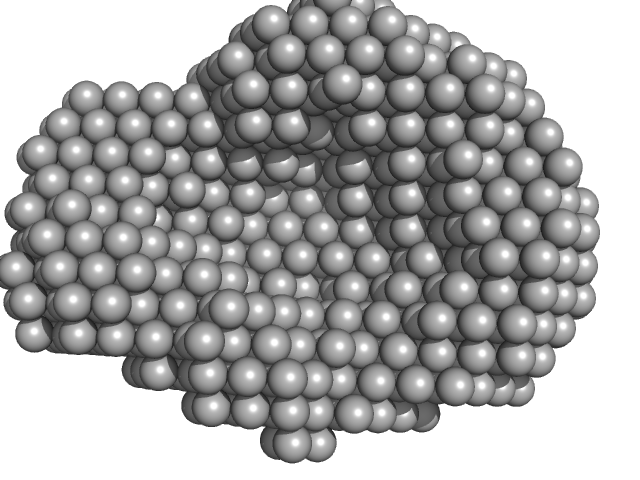
| Sample: |
Accessory colonization factor monomer, 121 kDa Escherichia coli (strain … protein
|
| Buffer: |
20 mM Tris, 200 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2021 Apr 21
|
Molecular and cellular insight into Escherichia coli SslE and its role during biofilm maturation
npj Biofilms and Microbiomes 8(1) (2022)
Corsini P, Wang S, Rehman S, Fenn K, Sagar A, Sirovica S, Cleaver L, Edwards-Gayle C, Mastroianni G, Dorgan B, Sewell L, Lynham S, Iuga D, Franks W, Jarvis J, Carpenter G, Curtis M, Bernadó P, Darbari V, Garnett J
|
| RgGuinier |
3.4 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
171 |
nm3 |
|
|